The Stability of the Bali’s Rabies Virus Molecular Marker for the Development of Diagnostic Method
on
Jurnal Ilmu dan Kesehatan Hewan, Agustus 2015
Vol 3 No 2: 45-50
The Stability of the Bali’s Rabies Virus Molecular Marker for the Development of Diagnostic Method
Stabilitas Penanda Molekuler Virus Rabies Bali untuk Pengembangan Metode Diagnosa
Putu Dimas Abiyoga1,2*, Gusti Ayu Yuniati Kencana1, I Nyoman Dibia2 1Magisterial Program of Veterinary Medicine, Udayana University Jl. PB Sudirman, Denpasar 2Laboratory of Virology, Animal Disease Investigation Centre, Denpasar Corresponding author: abiyogadimas@yahoo.co.id
ABSTRAK
Penelitian ini bertujuan untuk membuktikan penanda molekuler virus rabies Bali masih tetap conserve dan untuk pengembangan metode diagnosa rabies berbasis penanda molekuler. Tiga puluh sampel otak anjing yang terinfeksi rabies pada tahun 2014 dan 2015 digunakan pada penelitian. Sekuen nukleotida yang yang didapatkan dari penelitin dan yang diakses di GenBank dianalisis dengan program MEGA 5.2. Hasil analisis membuktikan bahwa asam amino spesifik Isoleusin pada posisi 308 (open reading frame) sebagai penanda molekuler virus rabies Bali masih conserve. Gen N yang diamplifikasi dengan teknik RT-PCR yang menggunakan primer spesifik menunjukkan bahwa masing-masing isolate virus rabies memiliki pita khas , dapat membedakan isolat Bali dengan isolat lain di Indonesia.
Kata kunci: Bali, rabies, penanda molekuler, diagnosa, RT-PCR.
ABSTRACT
The objectives of this study were to prove that the molecular marker of Bali’s rabies virus is still conserve and to develop a diagnostic method based on molecular marker. Thirty brain samples of dog that had been infected by a rabies virus from 2014 and 2015 were used for this research. The sequences of nucleotide which were obtained and the sequences of nucleotide accessed in GenBank were analyzed using MEGA 5.2 software. The result provided that the specific amino acid (isoleusin) at position 308 (open reading frame) as a molecular marker of Bali’s rabies virus was still conserve. Fragment of N gene amplified by Reverse RT-PCR method with a specifically designed primers showed that every isolate of rabies virus had it’s own typical band, and could distinguish Bali isolate from the others in Indonesia.
Keywords: Bali, rabies, molecular marker, diagnose, RT-PCR
INTRODUCTION
Based on the Decree of the Minister of Agriculture No. 4026/Kpts /OT.140/3/2013, rabies is grouped into Strategic Animal Diseases (PHMS) and given priority in the prevention, control and eradication. Cases of rabies in Bali confirmed in the laboratory first time in November 2008. The disease raises serious public health problem in all districts/municipalities in the Province of Bali. Efforts to control and eradicate rabies have been implemented over five years through a mass vaccination program in Bali, but until now the cases of rabies in animals (especially dogs) still occur. Outbreaks of
rabies occurred in Bali is very likely due to the inclusion of rabies infected animals.
Rabies is a fatal zoonotic disease that is caused by the rabies virus. As a member of the RNA virus, rabies virus is often characterized by excessive genetic variation by mutation during replication. This occurrence is due to the absence of a mechanism for proofreading and error correction after replication by RNA polymerase enzyme. The most common mutation occurred in rabies virus is a mutation of a single nucleotide in the genome of a virus called point mutations. The occurrence of point mutations often happen naturally (Ratam, 2005). Point mutations can
be synonymous or non-synonymous substitutions. In synonymous nucleotide substitution, nucleotide change is not followed by changing in the amino acids of the protein expressed. According to Nagarajan et al. (2006) changing to a new specific amino acids that occurred from non-synonymous substitutions in a gene of rabies virus can produce a new molecular marker. Dibia et al. (2014), stated that the rabies virus of Bali isolates has a molecular marker composed of amino acid isoleucine at position 308 (open reading frame). That molecular markers can be used as markers to investigate the dynamic and distribution of rabies virus epidemiologically. Therefore, the molecular characterization of the rabies virus isolates that keep in nature needs to be done. This research aimed to prove that the Bali rabies virus is still conserve throughout the years using a specific molecular marker.
MATERIALS AND METHODS
Thirty samples of brains of animals were used in this study. The samples were collected from dogs infected by rabies in 2014 and 2015 from several areas in Bali. For the comparation, the research also used each one brain of dog from Sumatra, Java, Kalimantan, Sulawesi and Flores that detected positive for rabies.
Flourescent Antibody Test (FAT)
Flourescent Antibody Test was done by making smears and fixed with acetone at 20°C for 30 minutes. After drying at room temperature, preparations were imbued with anti-rabies nucleocapsid conjugate. Incubation was done inside the incubator at 37 ° C for 30 minutes. Then, preparations were washed with a solution of PBS pH 7.2 for 3 times and followed by adding aqueous mounting media and closed with a cover slip. Furthermore, the examination was conducted under a fluorescent microscope and neuron cells infected with the rabies virus will be marked by a fluorescent green color (OIE, 2008; Trimarchi and Nadin-Davis, 2007; Direktorat Kesehatan Hewan, 2009).
RNA Isolation Rabies
Brains of animals that positive for the rabies virus based on FAT, were made into 20% suspension in PBS pH 7.4 and inactivated by SDS (450 mL of 20% brain suspension was added 50 mL of 10% SDS). RNA isolation was done by using Trizol®reagent (Invitrogen), according to the manufacturer's instructions. A total of 250 mL and 750 mL suspension brain Trizol incorporated into 1.5 ml eppendorf tube. Vorteks mixture for one minute. After incubation at room temperature (20-25 ° C) for 5 minutes, to the mixture was added chloroform 200 mL, then vortex 15 seconds and incubated at room temperature for 15 minutes. Next, the tube was centrifuged at a speed of 12,000 RCF for 15 minutes. Aquaeus part was taken and put into a sterile 1.5 ml eppendorf tube. It was added to 500 mL of isopropyl alcohol and mixed until homogeneous. The mixture was incubated for 10 min at room temperature. Furthermore eppendorf tube which already contains the mixture was centrifuged at 12,000 RCF for 10 min, the supernatant was discarded, and added alcohol 70% 1000 mL. After vortex and centrifuged at 7500 RCF for 5 min, the supernatant was discarded, while the sediment was dried at a temperature of 55°C in incubator and resuspended in ultrapure aquabidest (Otsu-WI, Otsuka). Subsequently, the RNA of the rabies virus was sequenced at Genetika Science, Jakarta.
Sequence Data Analysis
Data were analyzed using molecularbased MEGA 5.2 to determine the stability of specific amino acids as molecular markers of the Bali rabies virus. The nucleotide sequence obtained by this research was confirmed by the program of Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi. nlm.nih.gov/Blast.cgi) (Kumar et al., 2004) to identify the typical sequence of nucleoprotein gene fragment (N) of rabies virus. The sequence of nucleotides obtained in this study was then lined with other nucleotide sequences of the rabies virus in Indonesia and in various countries that were accessed from
the GenBank (http://www.ncbi.nlm.nih.gov/ genomes) using ClustalW alignment. GenBank access number of the rabies virus nucleotide sequences that were analyzed was displayed in analysis results. Determination of the presence of rabies virus genetic variation was based on the sequences of genes that encoding the nucleoprotein (N) and analyzed using MEGA 5.2 program (Tamura et al., 2007).
Development of RT-PCR Method
Thirty samples of brains of dogs that positive for rabies by DFAT were amplified using RT-PCR with a general and specific primers. On the reverse transcription (RT), complementary DNA (cDNA) ofthe rabies virus genome was synthesized using NF36 (C/T) (forward primer) (5'-TCAGGTGGTC TCYTTGAAGCC-3') position 36-56 nucleotides and NR1251 (reverse primer) (5'-TTAGTCGACCTCCGTTCA-3') positions 1232-1251 nucleotides. The primers is belong
to a general primer. Then, the primer pair NF898 (S) (forward primer) (5'-TACTCATCTAATGCAGTTGGTCACA-3 ') positions 898-922 nucleotides and NR1349 (reverse primer) (5'-CGAGTCACTAGAA TACGTCTTGTT-3') position 1326-1349 nucleotides, are a primary partner that specifically flank 451 base pairs.
RESULTS AND DISCUSSION
Results
Detection of rabies virus antigen using methods DFAT has been carried out on 30 samples of brains of dogs that were diagnosed rabies from various regions in Bali and the samples from Sumatra, Kalimantan, Sulawesi, Java, and Flores showed rabies positive. A specific aggregate (nucleocapsid protein) can be detected by luminescence apple green or greenish yellow with a dark background (Figure 1).
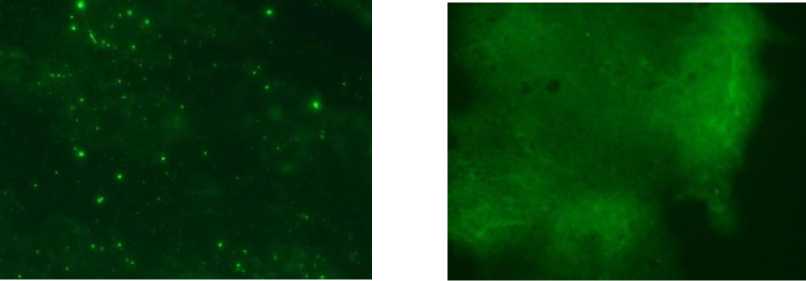
Samples were positive FAT shown in Figure 1 CA.). In winch brain samples are aggregates of nucleocapsid protein are apple green or greenish yellow with a Harle background Whereas Figure 1 (B) brain samples HiH not indicate the presence of nucleocapsiH protein are apple green or greenish yellow.
Molecular Marker Analysis
The amino acid sequences of the Bali rabies virus nucleoprortein had a typical amino acid at position 308. This typical amino acid was isoleucine, which was different from others, the valin amino acid (Figure 2). The amino acid sequences of the Bali rabies virus nucleoprortein were obtained from the translation of nucleotide sequences using MEGA software and the other sequences got from on line searching using the program of Basic Local Alignment Search Tool (BLAST)
(http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Kumar et al., 2004), and aligned by the program of ClustalW.
Development of RT-PCR
Thirty samples of brains of dogs that were positive for rabies by DFAT successfully amplified using RT-PCR with the general and specific primers. The general primer produced the 1251 bp of band length (Figure 3), while the specific primer yielded 451 bp of band length (Figure 4).
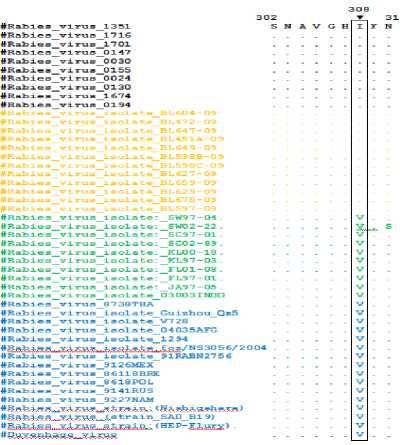
figured Alignment of amino acid sequences Bali rabies vims nucleoprotein and rabies viruses that are accessed on the GeneBank. The same amino acid is indicated by dots. Specific substitution at position 308 indicated by the arrow in the top of the box.
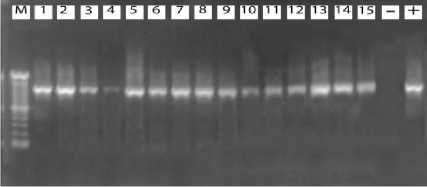
Figure. 3: Results of electrophoresis OfRT-PCRproducts7 M=marker (1 OObp ladder DNA) (+) =positive control, (-) =negative control, Lane 1-10= SampleBali rabies virus, Lane 11-15 = Samples viruses out Bali. The arrows indicate product-specific RT-PCR at position 1215 bp.

Eig⅛re,.⅜, Results of electrophoresis ofRT-PCRproducts, M=marker (IOObpladder DNA) (+) =positive control, (-)=negative control,Lane 1-10=samplesBali rabies virus, Lane 11-15 = Samples viruses out Bali. The arrows indicate product-specific RT-PCR at position 451 bp.
Discussion
Detection of Rabies Virus Using FAT
According to Dean et al. (1996);
Trimarchi and Nadin-Davis (2007) test results using the FAT has been in accordance with the standard requirements. The FAT is one of the test to detect rabies antigen in the network, with a sensitivity of 98-100% of
cases of rabies (McColl and Lunt., 2003; Tepsumethanon et al., 2004; OIE, 2008). The sensitivity of the test which is owned by FAT makes this test becomes a recommendation of World Health Organization as a standard test for rabies diagnose (OIE, 2008; Wacharapluesadee et al., 2008).
However, the sensitivity of the FAT test is influenced also by the quality of the samples, conjugate, equipment and capabilities of diagnostic tools (McElhinney et al., 2008). The test results which are efficient, fast and accurate are essential for the management of rabies carrier animal bitten patients (Akoso, 2007). But according to David et al. (2002), this technique has the disadvantage of not being able to detect viral antigens in brain specimens that had rotted. Besides, the FAT is unable to distinguish the specific strains of rabies virus. Therefore, RT-PCR can be used as a complementary diagnostic tool for the FAT testing, mainly for animal brain samples taken after 8 hours of death.
Stability of Molecular Markers of Bali Rabies Virus
Results of the analysis of amino acid sequences derived from the nucleoprotein gene showed that molecular markers of Bali rabies virus isolates is still conserve. Dibia et al. (2014) reported that Bali rabies virus has a substitution of non synonym unique amino acid isoleucine at position 308 (Open Reading Frame), while all of the rabies virus in Indonesia and in other countries in the world as well as some of the vaccine strains have amino acid valin in the same position (Figure 2).
Among the genes that are owned by the rabies virus, nucleoprotein gene is a gene that is highly conserve with 99% homology (Ito et al., 2001). Previous researchers, Konznetzoff et al. (1998) identified that the N gene regions that most conserve locate at amino acid position 298-352.
Mutational events that occur in viral isolates of rabies in Bali has shown its character as a member of RNA viruses (Dibia et al., 2014b). While, Murphy et al. (2007)
stated that RNA viruses are characterized by a high rate of mutation during replication in the absence of proofreading and repair mechanisms mistake after replication by RNA polymerase enzyme. This study confirms that the the nucleoprotein of Bali rabies virus has a typical amino acid, isoleucine, at position 308, , which is not owned by the isolates of rabies in Indonesia and a few countries in the world. In addition, specific amino acids (isoleucine) is the part that most conserve in the N genes of rabies virus, so it can be said that the isoleucine at position 308 (open reading frame) of the nucleoprotein gene is a molecular marker for Bali rabies virus and it can be used as epidemiological markers to investigate the dynamics and distribution of the virus.
Development of RT-PCR
The results of RT-PCR indicate that the primers used are specific and stick to the expected position. In addition, the primer that is specifically designed in this study is able to detect both the N gene of rabies virus in animals of some infected areas in Bali. These results also indicate that the RT-PCR that developed have a high agreement with FAT method, and is able to distinguish the rabies virus of Bali isolates with rabies virus isolated from outside Bali, so this test can be used as a reliable confirmatory diagnostic tool.
The results support a similar study conducted by Benedictis et al., (2011) which revealed the superiority of RT-PCR. Furthermore, Benedictis et al. (2011) reported that one-step RT-PCR showed relatively high specificity of 98.94% (CI: 97.55 to 99.65), the sensitivity of 99.71% (CI: 98.40 to 99.99) and accuracy 98.90% compared with the values obtained by DFAT used as the standard method of diagnose of rabies. Moreover, Benedictis et al., 2011 stated that the agreement between the test method of one step RT-PCR and the standard method of FAT has an almost perfect agreement with Cohen's Kappa coefficient of 0.977. In addition, the advantage of using RT-PCR is that the method is still able to detect viral antigens in brain specimens that had rotted
which was not detected using FAT (David et al., 2002).
CONCLUSION
-
1. Molecular markers of Bali rabies virus at position 308 (open reading frame)
remained stable.
-
2. The method of RT-PCR using specific primers of Bali rabies virus strain is able to distinguish between the Bali and non Bali rabies virus.
ACKNOWLEMENT
Author to convey gratitude to the Head of Center for Veterinary, Denpasar for giving research permission and facilities.
REFERENCES
Akoso BT. 2007. Pencegahan dan Pengendalian Rabies.Yogyakarta: Penerbit Kanisius.
Benedictics PD, Battisti CD, Dacheux L, Marciano S, Ormelli S, Salomoni A, Caenaszzo ST, Lapelletier A, Bourhy H, Capua I, Cattoli G. 2011. Lyssavirus Detection and Typing Using Pyrosequencing. J Clin Microbiol 49(5) : 1932-1938.
David D, Yakobson B, Rotenberg D, Dveres N, Davidson I, Stram Y. 2002. Rabies Virus Detection by RT-PCR in Decompoosed Naturally Infected Brains. Vet Microbiol 87(2) : 111-118.
Dean DJ, Abelseth MK, Anatasiu P. 1996. The Fluorescent Antibody Technique.In: Meslin, F. X., Kaplan, M. M., Koprowski, H., (ed). Laboratory techniques in rabies, 4th ed. WHO: 88-95.
Dibia N, Sumiarto B, Susetya H, Putra AAG, Mahardika IGNK, Scott-Orr H. 2014. Diagnosis and Molecular Marker Analysis of Bali Rabies Virus Isolates. J. Vet. Vol.15(3): 288-297.
Direktorat Kesehatan Hewan, 2009. Standar Nasional Metode Diagnosa Rabies. Jakarta: Direktorat Jenderal Peternakan, Departemen Pertanian. Jakarta.
Ito N, Kakemizu M, Ito KA, Yamamoto A, Yoshida Y, Sugiyama M, Minamoto N. 2001. A comparison of complete genom sequences of the attenuated RC-HL strain of rabies virus used for production of animal vaccine in Japan, and the parentral Nishigahara strain. Microbiol.Immunol. 45: 51-58.
Kumar S, Tamura K, Nei M. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetic Analysis and sequence alignment. Brief. Bioinfor. 5(2): 150-163.
Kouznetzoff A, Buckle M, Tordo N. 1998. Identification of a region of the rabies virus N protein involved indirect binding to the viral RNA. J Gen Virol 79 : 1005-1013.
McColl KA, Lunt RA. 2003. Australia and New Zealand Standard Diagnostic Procedures. Victoria: CSIRO Australian Animal Health Laboratory.
McElhinney LM, Fooks AR, Radford AD. 2008. Diagnostic tools for the detection of rabies virus.European Journal Of Companion Animal Practice 3: 224-231.
Murphy FA, Gibbs EPJ, Horzinek MC, Studdert MJ. 2007. Veterinary Virology.Third edition. USA: Elsevier, Academic Press.
Nagarajan T, Mohanasubramanian B, Seshagiri EV, Nagendrakumar SB, Saseendranath MR, Satyanarayana ML, Thiagarajan D, Rangarajan PN, Srinivasan VA, 2006.
Molecular Epidemiology of Rabies Virus Isolates in India.J. Clin. Microbiol. 44(9):
3218-3224.
OIE (Office International des Epizooties). 2008. Manual of Standards for Diagnostic tests and Vaccines. Chapter 2.1.13: 304-323.
Ratam FA. 2005. Virologi. Cetakan pertama. Surabaya: Airlangga University Press.
Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 42(8): 1596-1599.
Tepsumethanon V, Lumlertdacha B,
Mitmoonpitak C, Sitprija V, Meslin FX, and Wilde H. 2004. Survival of naturally infected rabid dogs and cats.Clin. Infect. Dis. 39: 278280.
Trimarchi CV., and Nadin-Davis SA., 2007. Diagnostic Evaluation. In: Jackson AC., and Wunner WH., (ed.). Rabies. Second edition. USA: Elsevier, Inc.: 411-470.
Wacharapluesadee S., Sutipanya J.,
Damrongwatanapokin S., Phumesin P., Chamnanpood P., Leowijuk C., and
Hemachudha T. 2008. Development of a TaqMan real-time RT-PCR assay for the detection of rabies virus.J. Virol. Methods 151: 317-320.
50
Discussion and feedback