IN SILICO PRIMER DESIGN AND ANNEALING TEMPERATURE OPTIMIZATION TO AMPLIFY THE FRAGMENT OF gyrB GENE Mycobacterium tuberculosis ISOLATE P010 USING POLYMERASE CHAIN REACTION
on
Journal of Health Sciences and Medicine, Vol. 2 No. 1 February 2018
IN SILICO PRIMER DESIGN AND ANNEALING TEMPERATURE OPTIMIZATION TO AMPLIFY THE FRAGMENT OF gyrB GENE Mycobacterium tuberculosis ISOLATE P010 USING POLYMERASE CHAIN REACTION
Putu Irma handayani1, Dyah Subadrika Warma Dewi1, Ni Putu Eka Nata Sari1, Sang Ayu Made Rai Wulan Dewi1, Sagung Chandra Yowani1,3, Ni Nyoman Ayu Dewi2,3, Putu Sanna Yustiantara1,3
1Department of Pharmacy, Faculty of Mathematics and Natural Science, Udayana University 2Department of Medicine, Udayana University
3MDR-TB & XDR-TB Research Group, Udayana University
Bali, Indonesia
E-mail : handayaniirma7@gmail.com
Abstract
One of the factors causing XDR-TB is due to mutations in the Mycobacterium tuberculosis gene, one of them is in the gyrB gene. Amplification of gyrB gene fragments from Mycobacterium tuberculosis DNA using Polymerase Chain Reaction (PCR) method. The amplification process by the PCR method requires a pair of primers (forward and reverse) to limit the area to be amplified. The current study aims to obtain the best primer pair generated by in silico design using Clone Manager Suite 6 program while simultaneously optimizing the annealing temperature to amplify the fragment of gyrB Mycobacterium tuberculosis. The template used in designing the primer is the sequence of gyrB Mycobacterium tuberculosis H37Rv isolate obtained from NCBI database of genbank code AL123456.3.
The current study obtained a pair of primer which respectively had 19 oligonucleotide length and the best annealing temperature of 56ºC. The primer is be able to do in silico amplification of the fragment of gyrB Mycobacterium tuberculosis gene isolate P010 in the nucleotide area range from 1271-1755 bp with 485 bp fragment length.
Key words: primer design, annealing temperature, gyrB gene, PCR.
-
I. INTRODUCTION
Indonesia is included in 30 countries with a high burden of MDR-TB which reaches 32,000 new cases of MDR-TB from 480,000 global data on new cases of MDR-TB [8]. According to WHO data (2017) MDR-TB cases have grown by 6.2% to Extensively Drug Resistant Tuberculosis (XDR-TB). XDR-TB is a TB caused by bacteria resistant to INH and RIF (MDR-TB) moreover it is also resistant to at least Fluorokuinolone (FQ) and one of the second-line tuberculosis drugs in the form of injection [8]. In an tuberculosis drugs resistance treatment study, an examination has been performed on some second-line tuberculosis drugs. It is found out that Fluorokuinolone (FQ) is the most important drug of
the core MDR-TB regimen, this is due to the effectiveness of the therapy given and the low toxicity of the use of FQ compared to other second-line OAT.
The mechanism of development of FQ resistance in M. tuberculosis is the presence of mutations in the Quinolone Resitance Determining Region (QRDR) gyrB gene [5]. QRDR gyrB gene is defined as the region at codons 500 to 540 [4].
The identification of mutations in gyrB gene fragments can be done by PCR technique. In the process of amplification of gene fragments required a pair of primers (forward and reverse) is required. Primer serves as a limiting fragment of DNA to be amplified. The primer used must meet the criteria of a
good primer to amplify gyrB fragments specifically. These criteria include: primary length,% GC, Tm, dimer and hairpins, primary stability, repeats, runs and false priming. Therefore the current study aims to design a pair of primer used to amplify gyrB gene fragment as well as to find out the optimum annealing temperature to amplify gyrB fragment by conducting annealing temperature optimization.
-
II. METHODS
-
2.1 Primer Design
-
Primer used to amplify gyrB gene fragment in the current study was designed to utilize Clone Manager Suite 6 program. The first stage to be done was inputting the sequence nucleotide data of gyrB M. tuberculosis data (Gen Bank AL123456.3), which was provided in the database at URL://www.ncbi.nlm.nih.gov, to Clone Manager Suite 6 program. A Pair of primer was designed by selecting menu bar primer design. The length of primer base produced was input and target area to amplified based on the mutation data literature was determined. Primers produced in the rank line was analyzed based on the primer criteria desired for gyrB gene area.
-
2.2 Annealing Temperature Optimization and Amplification of gyrB M. tuberculosis Gene Fragment using PCR
Determination of primer optimum temperature to be used to amplify gyrB fragment on isolate H37Rv was done by conducting an experiment at 56°C, 58°C, 60°C
DNA template used for amplification was obtained from the isolation result of isolate P010. PCR was conducted using a pair of primer specially designed on the current study used Clone Manager Suite 6 where the amplification process started with predenaturation at 95°C for 15 minutes followed by 45 amplification cycle (denaturation at 94°C for 1 minute, annealing at optimization temperature at 56°C, 58°C, 60°C for 1 minute 20 seconds, extension at 72°C for 2 minutes and final extension at 72°C for 10 minutes).
-
2.3 PCR Product Detection
PCR product was detected using electrophoresis agarose gel 1,5% b/v. The result of electrophoresis was then conducted visualization using UV Transilluminator.
-
III. RESULT AND DISCUSSION
-
3.1 Primer Design
-
The primer design performed using Clone Manager Suite 6 obtains the result of a pair of primer which are forward primer with a nucleotide base
sequence:5'-GAGAGTTGGTGCGGCGTAA-3' and reverse primer with a nucleotide base sequence:5'GCGGTCGGAGTATGCGAAT-3'.
The length of the forward and reverse primer of gyrB genes is 19 bases. Primers that have a length of less than 18 bases will have lower specificity and will easily cause mispriming [2].
Percent (%) of GC of forward and reverse primer of gyrB gene show the same amount that is 57%. The GC percentage has met the primer general requirements of 40% to 60% [7]. If a primer has a low GC level, the primer will not be able to attach effectively to the target so that there will be a decrease in the efficiency of the PCR process. In the other hand, the high level of GC will complic ate the separation of double helix chains on primers and templates [2].
Forward and reverse primers of the gyrB gene have the same Tm (melting temperature) of 64 °C. Tm is the temperature at which 50% of the DNA's double helix has been separated. Tm in a pair of primer of gyrB gene has met the criteria ranging from 50-65 ° C and do not have a gap of Tm between one primer with the others [2]. A pair of primer should not have a high gap of Tm. The gap recommended is not more than 5°C, if it is higher, it will cause a decrease in the amplification process or even the amplification process can be performed. It also aims to facilitate the determination of annealing temperatures in the multiplex-PCR process [7].
The pair of primer generated from the design has a dimer at the tip of 3’ as much as 2 pb on each forward and reverse primer, while dimer-any presence as much as 2 pb on forward primer and 3 pb on reverse primer. It shows that primer generated from the design still meets the criteria of Clone Manager suite 6 program which has a tolerance limits of dimer as much as <3 pb at the tip of 3’ and <7pb other than 3’ area.
The designed primer has a stability of 2.5 kcals and 2.3 kcals for forward and reverse primers respectively. So the stability of the two primer design has met the criteria of Clone Manager Suite 6 that is more than or equal to 1.2 kcals. This figure indicates the stability of 5 bases (pentamer) of the tip of 5 ' is greater than at the tip of 3'.
Runs and repeats that are allowed in the Clone Manager Suite 6 program are less than 3. In the primer generated from the design, both primers meet the allowed runs and repeats criteria. Repetition of both nucleotide and dinucleotide bases that is more than 3 will increase the possibility of false priming [6].
Hairpins condition was not found in the primer design so it has met the criteria of a good primer based on Clone Manager Suite 6. Hairpins is a
condition which the tip of primers complement each other [3].
The last important criterion is false priming. When the presence possibility of false priming is not found in the pair of primer generated from the design, the primer design has met the Clone Manager Suite 6 criteria. False priming is not allowed to occur on primers. The existence of false priming will result in an amplification error beyond the annealing temperature so that the desired PCR product is not formed [1].
Based on literature and criteria reference of Clone Manager Suite 6, it can be concluded that a pair of primer design for gyrB gene amplification has met the criteria of a good primer. Therefore, the primer can be used to amplify the gyrB gene sequence so that it is able to produce the desired PCR product.
Table 1. The result of Primer Design of gyrB gene
based on the reference in Clone Manager Suite 6
|
Criteria |
Primer For-ward |
Primer Reverse |
References to Clone Manager Suite 6 |
|
Long |
19 |
19 |
18-22 |
|
primer (basa) Percent |
57 |
57 |
50-60 |
|
(%) GC Tm (°C) |
64 |
64 |
55-80 |
|
Dimer on |
2 |
2 |
<3 |
|
tip 3’ Dimer |
2 |
3 |
<7 |
|
besides the tip 3’ Stabili-ty |
2.5 |
2.3 |
≥1,2 |
|
(kcals) Runs |
2 |
2 |
<3 |
|
(nukleoti da) Repeats |
2 |
- |
<3 |
|
(dinukleo tida) Hairpins |
- |
- |
- |
|
False |
- |
- |
- |
|
priming |
-
3.2 Optimization of Annealing Temperature and Amplification of the Fragment of gyrB M. Tuberculosis Gene Isolate P010 Using PCR Technique
Annealing temperature is temperature required for the primer to attach to the target DNA. The low
annealing temperature of the optimum temperature will result in false priming and if the annealing temperature used is higher than the optimum temperature, it will cause the primer to be unable to attach to the target DNA so that the expected PCR product will not be formed, in other words the PCR process is not successful [2]. Threfore, it is necessary to process annealing temperature optimization so that the process of PCR will work properly and produce the product which is appropriate to the target.
Optimization of PCR process on the annealing temperature in this study used 56ºC, 58 ºC dan 60 ºC. Temperature variations are performed with the aim of obtaining the most optimum temperature for the PCR process so that the amplification process will also work as it is planned. PCR in this study was conditioned at 95 ° C for 15 minutes for predenaturation, followed by 45 cycles of amplification with denaturation at 94 ºC for 1 minute, annealing temperature variation 56 ºC, 58 ºC dan 60 ºC for 1 minute 20 seconds, elongation at 72 ºC for 2 minutes and final elongation process at 72 ºC for 10 minutes. From annealing temperature optimization, it is obtained amplicon visualized by the process of electrophoresis agarose 1.5% w / v.
0,5 kb
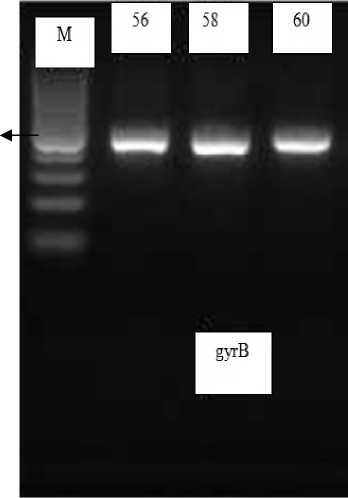
Figure 1. Electroforegram amplification gyrB gene fragment isolate P010 on UV- transluminator; gyrB= product gyrB gen; M= Marker DNA ladder 100 bp (0,1 kb)
3 bands with different optimization temperatures show the same result, just as bright and bold. The number of products produced is also
suitably seen on marker ±485 bp. If the resulting product is as good at two different temperatures, in this case there is no secondary hybridization and the DNA bands obtained are sufficiently thick, then a lower annealing temperature will be selected [1]. This consideration is caused by high annealing temperatures that can inhibit primary hybridization with DNA templates resulting in fewer PCR products so that temperatures 56 ºC can be used for gyrB gene amplification with primer design results. So that the PCR process is conducted at 56 ºC annealing temperature according to the Clone Manager Suite 6 annealing temperature criteria is 56 ºC.
After optimizing the annealing temperature and amplification using the PCR method on the P010 isolate using a pair of primer design with Clone Manager Suite 6 that is a forward primer with a nucleotide base sequence:
5'-GAGAGTTGGTGCGGCGTAA-3' and reverse primer with nucleotide base sequence:
5'-GCGGTCGGAGTATGCGAAT-3' proved to be able to amplify the gyrB gene in the P010 isolate used as a template.
It can be seen by detecting the PCR products performed using electrophoresis method on 1.5% agarose gel seen on UV Transilluminator. The electrophoresis results showed that PCR products formed correctly were products of ±485 bp in accordance with the size of the gyrB gene.
-
IV. CONCLUSION
In silico primer design produced primers which had good criteria as it had met the criteria such as primer length, %GC, Tm, dimer, hairpins, primer stability and false priming. A pair of primer obtained were primer forward: 5’-GAGAGTTGGTGCGGCGTAA-3’ and primer reverse:
5’-GAGAGTTGGTGCGGCGTAA-3’. It was proved that the pair of primer produced by in vitro were able to amplify gyrB gene fragment and produced amplicon with the size of ±485 bp after detecting using electrophoresis method and was seen using UV Transilluminator with temperature optimization results of 56 ºC.
ACKNOWLEDGEMENT
I would like to thank everyone who has helped me either directly or indirectly in order to conduct this study well.
REFERENCES
-
[1] Borah, P, 2011. Primer Designing for PCR. Science Vision. 11(3): 134-136.
-
[2] Handoyo, D. dan A. Rudiretna, 2000. Prinsip Umum Dan Pelaksanaan Polymerase Chain Reaction (PCR) (General Principles and Implementation Of Polymerase Chain Reaction). Unitas. 9 (1) : 17-29.
-
[3] Judelson, H. 2006. Guidelines For Designing Primers. Primer Guidelines. 10 (6) : 1-5.
-
[4] Li, J., X. Gao, T. Luo, J. Wu, G. Sun, Q, Liu, Y. Jiang, Y. Zhang, J. Mei and Q. Gao, 2014. Associaton of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerging Microbes and Infections.
-
[5] Palomino, J. C. and A. Martin. 2014. Drug Resistance Mechanisms in Mycobacterium tuberculosis. MDPI. Vol 3 : 317-340.
-
[6] Premier Biosoft International (PBI). 2009. Net Primer Manual. Available at:
http://www.PremierBiosoft.com. (Cited December 18, 2014).
-
[7] Sasmito, D. E.K., Rahadian, K., Izzati, Muhimmah. 2014. Karakteristik Primer pada Polymerase Chain Reaction (PCR) untuk Sekuensing DNA: Mini Review, Seminar Nasional Informatika Medis (SNIMed) V: Yogyakarta.
-
[8] World Health Organization (WHO). 2016. Global Tuberculosis Report 2016. Perancis: World Health Organization.
-
[9] World Health Organization (WHO). 2017. Global Tuberculosis Report. Available at:
http://www.who.int/tb/publications/global_report/ en/. (Cited December 20, 2017).
8
Discussion and feedback