COLON CANCER DRUG DEVELOPMENT STUDY OF ELAGIC ACID DERIVATIVES
on
p-ISSN 1907-9850
e-ISSN 2599-2740
JURNAL KIMIA (JOURNAL OF CHEMISTRY) 15 (2), JULI 2021 DOI: https://doi.org/10.24843/JCHEM.2021.v15.i02.p13
COLON CANCER DRUG DEVELOPMENT STUDY OF ELLAGIC ACID DERIVATIVES
L. R. Jannah and I G. M. Sanjaya*
Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Negeri Surabaya, Surabaya, Jawa Timur, Indonesia
*Email: igmasanjaya@unesa.ac.id
ABSTRAK
Penelitian ini bertujuan untuk mengembangkan obat kanker kolon dengan bahan baku senyawa asam Elagat dan turunannya menggunakan metode Hubungan Kuantitatif Struktur-aktivasi (HKSA) dan penambatan molekular. Deskriptor yang dipakai HKSA meliputi deskriptor elektronik, sterik, dan hidrofobik. Karakter dari masing-masing deskriptor dikomputasikan menggunakan software NWchem dan MarvinSketch dengan teori HF (Hartree-Fock). Penambatan molekul dilakukan dengan menggunakan software Autodock Tools dan Biovia. Hasilnya menunjukkan bahwa model persamaan terbaik HKSA senyawa bahan obat dari asam elagat dan turunanya adalah Log P = -7.103 + (-0.005*PSA) + (-0.729*MR) + (0.003*Volume) + (0.002*SA) + (-99.912*Homo) + (-38.893*Lumo)+ (-0.072*MD)+ (0.006*Eh) + (1.409*P) yang diperoleh untuk senyawa 2,3,7-trichloro-8-methoxychromeno[5,4,3-cde]chromene-5,10-dione. Hasil penambatan molekular tersebut memiliki energi ikat sebesar -9.94 kkal/mol dan konstanta inhibisinya 51,54 nM.
Kata kunci: asam elagat, HKSA, kanker kolon, penambatan molekular
ABSTRACT
This study aimed to develop a colon cancer drug with a raw material of ellagic acid and its derivatives using the Quantitative Structure-Activity Relationship (QSAR) method and molecular docking. The descriptors used in QSAR were electronic, steric, and hydrophobic descriptors. The characters of each descriptor were computed using NWchem and MarvinSketch software with the HF (Hartree-Fock ) theory. Molecular docking was performed using Autodock Tools and Biovia software. The results show that the best equation of QSAR model for medicinal compounds from ellagic acid and derivatives was Log P = -7.103 + (-0.005 * PSA) + (-0.729 * MR) + (0.003 * Volume) + (0.002 * SA) + (- 99,912 * Homo) + (38,893 * Lumo) + (-0.072 * MD) + (0.006 * Eh) + (1.409 * P) obtained for 2,3,7-trichloro-8-methoxychromeno [5,4, 3-cde] chromene-5,10-dione compound. The result of molecular docking had a binding energy of -9.94 kcal/mol and an inhibition constant of 51.54 nM.
Keywords: colon cncer, ellagic acid, molecular docking, QSAR.
INTRODUCTION
Cancer of the colon or colorectal cancer (CRC) is a disease affecting the colon and rectum. Colon cancer ranks third in the world and the cause of death from cancer(Ceci et al., 2018). At the beginning of 2020, approximately 147 950 people are diagnosed with colon cancer and 53 200 died of the disease, including 17 930 cases and 3,640
deaths in individuals aged under 50 years (Siegel et al., 2020).
One of the causes of colon cancer is the uncontrolled growth of Leukotriene A4 hydrolase (LTA4H), an enzyme that acts as an epoxide hydrolase and an aminopeptidase (Noviardi and Fachrurrazie, 2015). LTA4H can cause carcinogenesis(Zhao et al., 2019) and form leucotreine B4, which has a role in inflammation and cancer development (Vo, Jang and Jeong, 2018). LTA4H can be used as
an inflammatory marker in colorectal cancer. Therefore inhibition of LTA4H may decrease the development of colon cancer (Mira et al., 2020). Colon cancer treatment by chemotherapy, surgery, and radiation has not been effective in healing cancer. Therefore, compounds obtained from plants have attracted attention lately as an alternative for cancer prevention and treatment. The compounds from plants have low toxicity, cost, and side effects.
Ellagic acid is a natural polyphenol compound that has anticancer and antioxidant properties (Ceci et al., 2018). The anticancer activity present in the ellagic acid compounds can be used for various types of cancer (Wang et al., 2019). Ellagic acid and some of its derivatives can inhibit the proliferation of cancer cells, induce cell cycle arrest, and modulate some of the important cellular processes associated with cancer. (Molina et al., 2015). Ellagic acid can effectively inhibit the viability of colon cancer cells (HT29 and HCT 116) (Fang et al., 2015). Ellagic acid also have potential as inhibitors for epoxide hydrolase with IC50 values of 15.4 ± 1.3 μM (Luyen et al., 2015).
Based on this research, it is necessary to conduct a new drug development study by analyzing the QSAR and molecular docking. QSAR equation model, to predict the activity of ellagic acid and its derivatives as colonic anticancer compounds, was performed by multiple linear regression analysis. QSAR model is used for prediction and interpretation,
so it can help to understand the mechanism of the molecule used as the target. Chemical descriptors in QSAR modeling are used to describe various levels of chemical, physical, and structural characteristics of a molecule or target (Cherkasov et al., 2014)
Determination of molecular docking using the Discovery Studio Vistualizier (DSV) program can provide visualization of ligand interactions with a target molecule, so that it appears that amino acids play a role in maintaining the stability of ellagic acid derivatives against colon cancer protein, leukotriene A4 hydrolase (LTA4H). Testing the activity of ellagic acid derivatives against colon cancer with LTA4H colon cancer protein was carried out in silico with the method of quantitative relationship to structure and activity as well as the binding of LTA4H colon cancer protein with the code PDB: 3U9W.
MATERIAL AND METHOD
Material
The compound used in this study is ellagic acid and its derivatives are shown in Table 1. Ellagic acid derivatives are the result of functional group substitution expected to reduce the reactivity of ellagic acid compounds as anti-colon cancer. Ellagic acid has an LD50 range between 500-5000 with a mild toxic classification (BPOM, 2014).
Table 1. Compounds of ellagic acid and their derivatives
|
No |
Structure Name |
LD50 (mg/Kg) |
|
1. |
Ellagic Acid |
1712 |
|
2 |
3-chloro-2,7,8-trihydroxychromeno[5,4,3-cde]chromene-5,10-dione |
931,6 |
|
3 |
2-chloro-3,7,8-trihydroxychromeno[5,4,3-cde]chromene-5,10-dione |
849,3 |
|
4 |
2,7-dichloro-3,8-dihydroxychromeno[5,4,3-cde]chromene-5,10-dione |
1158 |
|
5 |
2,8-dichloro-3,7-dihydroxychromeno[5,4,3-cde]chromene-5,10-dione |
719,9 |
|
6 |
2,3,7-trichloro-8-hydroxychromeno[5,4,3-cde]chromene-5,10-dione |
1030 |
|
7 |
2,3,8-trichloro-7-hydroxychromeno[5,4,3-cde]chromene-5,10-dione |
1170 |
|
8 |
2,3,7,8-tetrachlorochromeno[5,4,3-cde]chromene-5,10-dione |
2360 |
|
9 |
2,3,7-trihydroxy-8-methoxychromeno[5,4,3-cde]chromene-5,10-dione |
2039 |
|
10 |
2,3,8-trihydroxy-7-methoxychromeno[5,4,3-cde]chromene-5,10-dione |
2092 |
|
11 |
3,8-dihydroxy-2,7-dimethoxychromeno[5,4,3-cde]chromene-5,10-dione |
2349 |
|
12 |
2,8-dihydroxy-3,7-dimethoxychromeno[5,4,3-cde]chromene-5,10-dione |
2032 |
|
13 |
3-hydroxy-2,7,8-trimethoxychromeno[5,4,3-cde]chromene-5,10-dione |
1599 |
|
14 |
2,7,8-trihydroxy-5,10-dioxo-5,10-dihydrochromeno[5,4,3-cde]chromene-3- |
1458 |
|
carboxylic acid |
No Structure Name
LD50
(mg/Kg)
15 8-chloro-2,7-dihydroxy-5,10-dioxo-5,10-dihydrochromeno[5,4,3-cde]chromene-3- 1533
carboxylic acid
16 2-chloro-7,8-dihydroxy-3-methoxychromeno[5,4,3-cde]chromene-5,10-dione1659
17 2,7-dichloro-3,8-dimethoxychromeno[5,4,3-cde]chromene-5,10-dione1569
18 2,8-dichloro-3,7-dimethoxychromeno[5,4,3-cde]chromene-5,10-dione1179
19 2,3,7-trichloro-8-methoxychromeno[5,4,3-cde]chromene-5,10-dione861,2
20 2-chloro-3,7,8-trimethoxychromeno[5,4,3-cde]chromene-5,10-dione1384
Methods
QSAR Method
The research used a theoretical experiment using the Quantitative StructureActivity Relationship (QSAR). The chemical structure of ellagic acid and its derivatives was drawn using Avogadro 1.2.0. The calculation of electronic descriptors using NWChem with the HF (Hartree-Fock) theory based on 631G* and steric and hydrophobic descriptors using Marvin Sketch. Multiple linear regression analysis using the Backward method was performed using the statistical software program SPSS version 16.0. Statistical analysis was applied to determine QSAR the equation model of ellagic acid compounds and derivatives. Table 2 shows the parameters used in QSAR method.
Table 2. QSAR parameters
|
Parameter |
Symbol |
Description |
|
Hydrophobic |
Log P |
Partition coefficient |
|
Steric |
SA MR V PSA P |
Surface Area Molar Refractifity Volume Polar surface area Polarizability |
|
Electronic |
Lumo Homo Δ Lumo-Homo DM H |
Lumo energy Homo energy The difference between lumo and homo energy Moment dipol Hydration energy |
Molecular Docking
Optimization process in molecular docking studies used Autodock Tools 1.5.6 software. Optimization of the target structure for colon cancer with the code PDB: 3U9W included adding hydrogen atoms and kollman.
The optimization results were stored in .pdbqt format, then the optimization results of the ellagic acid ligand and their derivatives were stored in .pdbqt format with a grid box setting of 60x60x60 Å to determine the location of the target mooring with the ligand, then the lowest affinity value was calculated. Amino acid determination at the active site was used to analyze the docking evaluation results using the Biovia Discovery Studio 2019 software.
RESULTS AND DISCUSSION
Ellagic acid compounds can induce apoptosis in various colon cancer cell lines with different mechanisms (Montane et al., 2020). This study using the QSAR method aimed to determine the correlation between biological activity and its properties. Physically, the parameters used were divided into three types including steric, hydrophobic and electronic. The biological properties of a molecule had many predetermined physicochemical parameters. The parameters described the biological activity used as the dependent variable and the physicochemical properties as the independent variable (Kapoor and Kumar, 2019).
Molecular descriptors were calculated using twenty compounds of ellagic acid and its derivatives which function as independent variables, while the observed inhibitory action (Log P) on cancer cells was the dependent variable with the QSAR model using multiple linear regression. Physicochemical properties are important properties of a molecule that affect the efficacy, safety, metabolism, and can predict using Lipinski's five rule, namely molecular mass <500, hydrogen bond donor <5, hydrogen bond acceptor <10, and LogP <5 (Lagorce et al., 2017). Ellagic acid and its derivatives met Lipinski's rule. The molecular descriptor used to model the QSAR equation is shown in Table 3.
JURNAL KIMIA (JOURNAL OF CHEMISTRY) 15 (2), JULI 2021 DOI: https://doi.org/10.24843/JCHEM.2021.v15.i02.p13
Table 3. Electronic, hydrophobic and steric descriptors
|
ID |
Log P |
PSA |
MR |
V |
SA |
Homo |
Lumo |
Δ Lumo-Homo |
DM |
H |
P |
|
1 |
1.16 |
133.52 |
70.61 |
219.86 |
319.91 |
-0.2004 |
-0.0301 |
0.1703 |
2.8153 |
29.87 |
26.99 |
|
2 |
1.31 |
113.29 |
73.43 |
256.57 |
369.63 |
-0.1991 |
-0.0337 |
0.1654 |
2.1839 |
27.09 |
28.29 |
|
3 |
1.96 |
113.29 |
73.43 |
258.34 |
372.63 |
-0.2049 |
-0.0291 |
0.1758 |
2.3317 |
24.34 |
28.56 |
|
4 |
2.76 |
93.06 |
76.26 |
260.91 |
372.97 |
-0.2113 |
-0.0293 |
0.1820 |
0.4431 |
20.47 |
29.84 |
|
5 |
2.11 |
93.06 |
76.26 |
262.70 |
377.06 |
-0.2050 |
-0.0339 |
0.1711 |
1.8496 |
39.34 |
29.86 |
|
6 |
2.92 |
72.83 |
79.08 |
261.71 |
370.83 |
-0.2109 |
-0.0346 |
0.1763 |
2.0417 |
16.79 |
31.44 |
|
7 |
2.27 |
72.83 |
79.08 |
263.51 |
375.03 |
-0.2047 |
-0.0380 |
0.1667 |
2.2654 |
36.34 |
31.18 |
|
8 |
3.07 |
52.60 |
81.90 |
264.30 |
372.49 |
-0.2104 |
-0.0387 |
0.1717 |
0.2293 |
29.50 |
32.76 |
|
9 |
0.54 |
122.52 |
75.09 |
271.48 |
356.75 |
-0.1982 |
-0.0286 |
0.1695 |
1.1093 |
28.09 |
28.88 |
|
10 |
1.19 |
122.52 |
75.09 |
273.26 |
356.86 |
-0.1992 |
-0.0287 |
0.1705 |
0.9530 |
25.32 |
29.14 |
|
11 |
1.22 |
111.52 |
79.57 |
290.76 |
393.77 |
-0.1997 |
-0.0289 |
0.1709 |
0.4041 |
22.53 |
31.30 |
|
12 |
0.57 |
111.52 |
79.57 |
290.09 |
393.89 |
-0.1987 |
-0.0286 |
0.1701 |
1.0804 |
24.39 |
31.04 |
|
13 |
0.6 |
100.52 |
84.05 |
308.27 |
430.70 |
-0.1992 |
-0.0288 |
0.1704 |
1.0892 |
21.43 |
33.20 |
|
14 |
0.96 |
150.59 |
75.88 |
275.22 |
345.85 |
-0.1987 |
-0.0559 |
0.1428 |
2.5584 |
30.78 |
28.96 |
|
15 |
1.11 |
130.36 |
78.71 |
277.81 |
350.54 |
-0.1985 |
-0.0570 |
0.1415 |
3.0007 |
43.84 |
30.26 |
|
16 |
1.34 |
102.29 |
77.91 |
272.70 |
361.73 |
-0.2041 |
-0.0288 |
0.1753 |
2.8987 |
23.41 |
30.45 |
|
17 |
1.53 |
71.06 |
85.52 |
294.14 |
403.58 |
-0.2083 |
-0.0289 |
0.1793 |
0.8162 |
31.94 |
33.90 |
|
18 |
1.53 |
71.06 |
85.22 |
294.14 |
403.38 |
-0.2048 |
-0.0339 |
0.1709 |
3.0767 |
34.45 |
33.92 |
|
19 |
2.3 |
61.83 |
83.56 |
279.22 |
371.41 |
-0.2094 |
-0.0342 |
0.1752 |
3.1339 |
34.91 |
33.33 |
|
20 |
0.76 |
80.29 |
86.88 |
309.06 |
435.72 |
-0.2038 |
-0.0289 |
0.1750 |
3.1903 |
19.37 |
34.50 |
The equation for the QSAR model used QSAR analysis of the Log P value with ten descriptors with the analysis method multiple linear regression carried out using the applied multiple linear regression. Multiple backward method. Statistical processing in this linear regression is a statistical technique used study resulted in 5 equation models.
in developing models (Ibrahim et al., 2018).
Table 4. Statistical parameter data of equation models
|
Model |
Variable |
R |
R2 |
Adj R2 |
SEE |
Significance |
F |
Ftable |
|
P, DM, Eh, Homo, | ||||||||
|
1 |
Lumo, SA, V, PSA, dan MR P, DM, Eh, Homo, |
0,996 |
0,991 |
0,984 |
0.10043 |
0 |
128,362 |
6,11 |
|
2 |
Lumo, SA, PSA, dan MR |
0,996 |
0,991 |
0,985 |
0,09767 |
0 |
152,628 |
5,86 |
|
Model |
Variable |
R |
R2 |
Adj R2 |
SEE |
Significance |
F |
Ftable |
|
P, DM, Eh, | ||||||||
|
3 |
Homo, Lumo, SA, V, dan MR |
0.995 |
0,990 |
0,985 |
0,09699 |
0 |
176,770 |
5,75 |
|
4 |
P, DM, Eh, Homo, Lumo, V, PSA, dan MR |
0,994 |
0,989 |
0,984 |
0,10094 |
0 |
190,107 |
5,79 |
|
5 |
P, DM, Homo, Lumo, SA, V, PSA, dan MR |
0,993 |
0,987 |
0,982 |
0,10584 |
0 |
207,033 |
5,99 |
In Table 4, the equation models 1-5 meet the requirements because Ftable has a value > 1 with a 95% truth level and an r2 value of more than 0.9. Equation model 1 is the best model with r2 of 0.9911 with the nine descriptors required as follows:
Log P = -7.103 + (-0.005*PSA) + (-0.729*MR) + (0.003*Volume) + (0.002*SA) + (-99.912*Homo) + (-38.893*Lumo)+ (-0.072*MD)+ (0.006*Eh) + (1.409*P).
Graph Model Equation 1
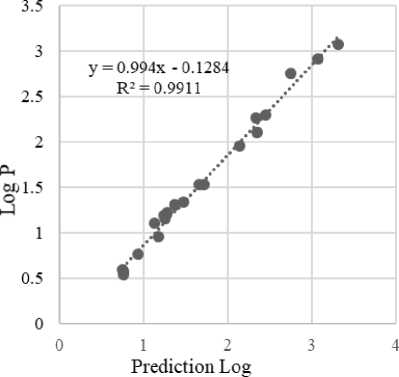
Figure 1. Graph Model Equation 1
Log P represents the hydrophobicity of a molecule. The more positive the log P value, the greater the lipophilicity. The greater
the P value, the more interactions the compound will have with the lipid phase (membrane)(Kapoor and Kumar, 2019). The compounds tend to be in a non-polar phase rather than a polar phase that causes compounds to penetrate easily the membrane and can bind to receptors. According to the log P parameter, ellagic acid is hydrophobic because log P is positive, so it has good bond selectivity to target proteins and is not toxic (Suciati, Lestari and Lukiati, 2020) (Syahputra, Ambarsari and Sumaryada, 2014). Based on the correlation analysis, it can be seen that the dipole moment descriptors, E Homo, E Lumo, polarizability, hydration energy and Log P showed a very close correlation. This is indicated by the regression value of 0.9911. In addition, the low Delta Lumo-Homo value indicates the compound has high reactivity, Homo has electrons which tend to give electrons as electron donors, while Lumo contains free places to accept electrons (Kumer, Sarker and Paul, 2019).
Molecular docking provides information on the interactions between compounds such as drugs or called ligands and enzymes or receptors through the active position in the enzyme by calculating the binding energy. Molecular docking calculations to predict the strength of the interaction between ligands and receptors (Sethi, Joshi and Sasikala, 2020). The calculation of the interaction energy in the form of a score docking (Ibrahim et al., 2018).
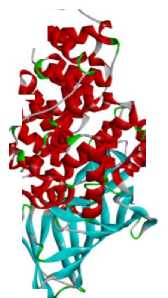
|
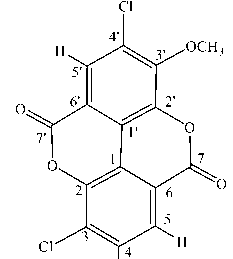
Molecular docking between Leukotriene A4 hydrolase and ellagic acid and its derivatives in Table 1.The following is the structure of the protein Leukotriene A4 hydrolase: ^^^” Figure 2. Protein structure of Leukotriene A4 Hydrolase (Source: https://www.rcsb.org/structure/3U9W) Tethering molecular compounds of ellagic acid was done to calculate the binding free energy (Gibbs energy, ΔG) of the ligand to the receptor protein and inhibition constant. Gibbs energy is an indicator to determine the stability of the interaction between ligands and receptors, so it can explain binding energies with different binding conformations. (IA, SO and OO, 2016). The inhibition constant was obtained from the binding energy (ΔG) using the formula Ki = eΔG/RT, R = 1.986 kcal/mol K, T = 298 K (Cheshomi, Reza and Matin, 2020)(Rowaiye et al., 2020). The lower the binding energy value, the smaller the inhibition constant value. Cl 7 1 I Cl'^^Yj^H Cl Figure 3. 2,3,7-trichloro-8-methoxychromeno[5,4,3-cde]chromene-5,10-dione Compound |
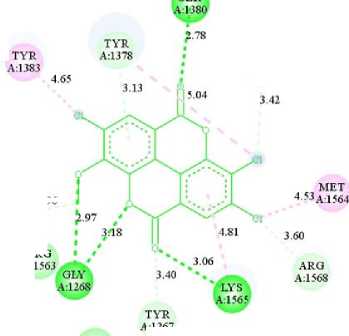
Figure 3 shows the structure of the molecule if the OH group on the C atom numbers 3, 4, and 4' that had been replaced with Cl and the C atom number 3' with OCH3 which caused the binding energy and the inhibition constant to decrease. The lower the binding energy caused the compound to be stable and could become colonic anticancer compounds. The results obtained from molecular docking were the binding energy of -9.94 kcal/mol and the inhibition constant of 51.54 nM. SER 2.78 TYR ; TYR A:1378 ∙ ≡ER A:1383 A:1379 4.65 343 ’« 3.42 MET 2.96 i I 1 453A1564 A:1269 * ; JΛ8 4-81 j60 AKO „ 563 ARQ θLY 3’4° A1568 A:1268 LYS A:1565 TYR A:1267 PRO A1266 Interactions CZl van der Waals ∏ Pi-Donor Hydrogen Bond ■1 Conventional Hydrogen Bond i ] Alkyl EZl Carbon Hydrogen Bond IB Pi-Alkyl Figure 4. Visualization of Amino Acid Interaction between Ellagic Acid and Leukotriene A4 Hydrolase Protein by Biovia Discovery Studio 2019 Software Ellagic acid compounds that interact via hydrogen bonds with the residue of Leukotriene A4 Hydrolase in colon cancer is the amino acid Gly1268 which bonded to the OCH3 group (2.97 Å), Gly1268 bonded to the ether group (3.18 Å), Lys 1565 bonded to the ketone group (3 , 06 Å) and Ser1380 bond to the ketone group (2.78 Å). CONCLUSION The QSAR equation model is shown with the results: Log P = -7.103 +(- 0.005*PSA) + (-0.729*MR) + (0.003*Volume) + (0.002*SA) + (- |
99.912*Homo) + (-38.893*Lumo)+ (-
0.072*MD)+ (0.006*Eh) + (1.409*P).
Molecular docking of ellagic acid and its derivatives obtained the lowest binding energy value of -9.94 kcal/mol and an inhibition constant of 51.54 nM in the 2,3,7-trichloro-8-methoxychromeno 5,4,3-cde] chromene-5 compound , 10-dione, so that it can inhibit the protein Leukotriene A4 Hydrolase in colon cancer.
REFERENCES
BPOM. 2014. Peraturan Kepala Badan Pengawas Obat Dan Makanan Republik Indonesia Nomor 2 Tahun 2014 Tentang Organisasi dan Tata Kerja Unit Pelaksana Teknis di Lingkungan BPOM, (875): 1– 111.
Ceci, C., Lacal, P., Tentori, L., De Martino, M., Miano, R., & Graziani, G. 2018. Experimental Evidence of the Antitumor, Antimetastatic and Antiangiogenic Activity of Ellagic Acid. Nutrients. 10(11): 1756. doi: 10.3390/nu10111756.
Cherkasov, A., Muratov, E. N., Fourches, D., Varnek, A., Baskin, I. I., Cronin, M., Dearden, J., Gramatica, P., Martin, Y. C., Todeschini, R., Consonni, V., Kuz'min, V. E., Cramer, R., Benigni, R., Yang, C., Rathman, J., Terfloth, L., Gasteiger, J., Richard, A., Tropsha, A. 2014. QSAR Modeling: Where Have You Been? Where Are You Going To?. Journal of Medicinal Chemistry. 57(12): 4977–5010. doi: 10.1021/jm4004285.
Cheshomi, H., Reza, A., Matin, M. M. 2020. Ellagic acid and human cancers: a systems pharmacology and docking study to identify principal hub genes and main mechanisms of action Database of Interacting Proteins. Molecular Diversity. 10:1007/ s11030-020-10101-6.
Fang, Y., Zhou, H., Xia, J. F., Lin, J. J., Li, R. Z., Yang, D. Q., Xu, M. Y., Li, X. Y. 2015. Ellagic acid regulates Wnt/β-catenin signaling pathway and CDK8 in HCT 116 and HT 29 colon cancer cells. Bangladesh J Pharmacol. 47–56. doi:
10.3329/bjp.v10i1.21068.
IA, A., SO, W. and OO, A. 2016. Molecular Docking Studies of Lonchocarpus cyanescens Triterpenoids as Inhibitors for Malaria. 6(2): 2–5. doi: 10.4172/21610398.1000213.
Ibrahim, Omotayo, A., Oyebamiji A. K., Oyewole, O. R., Semire, B. 2018. A DFT-Based QSAR and Molecular Docking Studies on Potent Anti- Colon Cancer Activity of Pyrazole Derivatives ADFT-Based QSAR and Molecular Docking Studieson Potent Anti-Colon Cancer Activity of Pyrazole Derivatives Strictly as per the compliance and regulations of. 18(2): 9-21.
Kapoor, Y. and Kumar, K. 2019. Quantitative Structure Activity Relationship in Drug Design: An Overview. SF Journal oof Pharmaceutical and Analytical
Chemistry. 2(2): 1–13.
Kumer, A., Sarker, N. and Paul, S. 2019. The theoretical investigation of HOMO, LUMO, thermophysical properties and QSAR study of some aromatic carboxylic acids using HyperChem programming. 3(1): 26–37. doi: 10.32571/ijct.478179.
Lagorce, D., Douguet, D., Miteva, M. A., & Villoutreix, B. O. 2017. Computational analysis of calculated physicochemical and ADMET properties of proteinprotein interaction inhibitors. Scientific Reports. 7(1): 1–15. doi:
10.1038/srep46277.
Luyen, B. T. T., Thao, N. P., Dat, L. D., Eun, K. J., Yang, S. Y., Kwon, S. U. Lee, Y. M., Kim, Y. H. 2015. Soluble Epoxide Hydrolase Inhibitory Activity from Euphorbia supina Rafin. Natural Product Science. 21(3): 176–184.
Mira, A., Sabry, M. A., Shimizu, K., & Abdel Bar, F. M. 2020. A new pimarane-type diterpene obtained by biotransformation inhibits human HCT-116 colorectal carcinoma through inhibition of LTA 4 H activity. Medicinal Chemistry Research. doi: 10.1007/s00044-020-02520-9.
Molina, A. R., Vargas, T., Molina, S., Sanchez, J., Martinez-Romero, J., Gonzalez-Vallinas, M., Martín-
Hernández, R., Sánchez-Martínez, R., Cedrón, M. G., Dávalos, A., Calani, L., Rio, D. D., González-Sarrías, A., Espín, J. C., Francisco A Tomás-Barberán, F. A., Reglero, G. 2015. The Ellagic Acid Derivative 4,4' -di- O -methylellagic Acid Efficiently Inhibits Colon Cancer Cell Growth through a Mechanism Involving WNT16’, Ramirez de Molina, A., Vargas, T., Molina, S., Sanchez, J., Martinez-Romero, J., Gonzalez-Vallinas, M., …
Reglero, G. (2015). The Ellagic Acid Derivative 4,4’-Di-O-Methylellagic Acid Efficiently Inhibits Colon Cancer Cell Growth through a Mechanism Involving WNT16. Journal of Pharmacology and Experimental Therapeutics. 353(2), 433– 444. doi:10.1124/jpet.114.221796
Montané, X., Kowalczyk, O., Reig-Vano, B., Bajek, A., Roszkowski, K., Tomczyk, R., Pawliszak, W., Giamberini, M., Mocek-Płóciniak, A., Tylkowski, B. 2020. Current Perspectives of the Applications of Polyphenols and Flavonoids in Cancer Therapy. Molecules. 25(15): 3342.
doi:10.3390/molecules25153342
Noviardi, H. and Fachrurrazie. 2015. Potensi Senyawa Bullatalisin Sebagai Inhibitor Protein Leukotrien A4 Hidrolase Pada Kanker Kolon Secara In Silico. 5(2): 65– 73.
Rowaiye, A. B Onuh, O. A., Sunday, R. M., Abdulmalik, Z. D., Bur, D., Emeter, N. W., Oluwaseun Adeola Obideyi, O. A., Pelletri, C. D., UjahSamuel, I. R., Iwuozor, C. R., Aondona, P. Y., Etalong, V. O., Yussuff, N., Akpa, J. N. 2020. Structure-Based Virtual Screening And Molecular Dynamic Simulation Studies Of The Natural Inhibitors Of Sars-Cov-2 Main Protease’, J Ong Chem Res, 5(1): 20–31. doi: 10.5281/zenodo.3767102.
Sethi, A., Joshi, K. and Sasikala, K. 2020. Molecular Docking in Modern Drug Discovery: Principles and Recent
Applications. Drug Discovery and Development - New Advances Empirical. 1–21. doi: 10.5772/intechopen.85991.
Siegel, R. L., Miller, K. D., Sauer, A. G., Fedewa, S. A., Butterly, L. F., Anderson, J. C., Cercek, A., Smith, R. A., and Jemal, A. 2020. Colorectal Cancer Statistics,
2020. 70(3): 145–164. doi:
10.3322/caac.21601.
Suciati, L., Lestari, S. R. and Lukiati, B. 2020. Molecular docking studies of geraniin, corilagin, and ellagic acid from rambutan (Nephelium lappaceum L.) peel extract against squalene synthase as potential anti hypercholesterolemia. Proceedings Of The 3rd International Seminar On Metallurgy And Materials (ISMM2019): Exploring New Innovation in Metallurgy and Materials. doi: 10.1063/5.0002534.
Syahputra, G., Ambarsari, L. and Sumaryada, T. 2014. Simulasi Docking Kurkumin Enol, Bisdemetoksikurkumin Dan Analognya Sebagai Inhibitor Enzim12-Lipoksigenase. Jurnal Biofisika. 10(1):
55–67.
Vo, T. T. L., Jang, W. and Jeong, C. 2018. Leukotriene A4 hydrolase: an emerging target of natural products for cancer chemoprevention and chemotherapy. doi:10.1111/nyas.13929.
Wang, Y., Ren, F., Li, B., Song, Z., Chen, P., & Ouyang, L. 2019. Ellagic acid exerts antitumor effects via the PI3K signaling pathway in endometrial cancer. Journal of Cancer. 10(15): 3303–3314.
doi:10.7150/jca.29738
Zhao, S., Yao, K., Li, D., Liu, K.,Jin, G.,Yan, M., Wu, Q., Chen, H., Shin, S. H., Bai, R., Wang, G., Bode, A. M., Dong, Z., Guo, Z., Dong, Z.Zhao, S., Yao, K., Li, D., Liu, K.,Jin, G.,Yan, M., Wu, Q., Chen, H., Shin, S. H., Bai, R., Wang, G., Bode, A. M., Dong, Z., Guo, Z., Dong, Z. 2019. Inhibition of LTA4H by bestatin in human and mouse colorectal cancer. EBioMedicine. doi: 10.1016/
j.ebiom.2019.05.008.
222
Discussion and feedback