DNA POLYMORPHISM AND GENETIC DIVERSITY OF MANGO (Mangifera sp.) GERMPLASM IN TROPICAL ISLAND
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 7 No. 1 • September 2019
eISSN: 2655-9994
pISSN: 2303-3371
https://doi.org/10.24843/IJBB.2019.v07.i01.p05
DNA POLYMORPHISM AND GENETIC DIVERSITY OF MANGO (Mangifera sp.) GERMPLASM IN TROPICAL ISLAND
Putu Suwardike1*, I Nyoman Rai1, Rindang Dwiyani1, and Eniek Kriswiyanti2
1Faculty of Agriculture, Udayana University, Jl. P.B. Sudirman, Denpasar, Bali, Indonesia 2Faculty of Mathematics and Natural Sciences, Udayana University, Jl. Raya Kampus Unud, Bukit Jimbaran, Badung, Bali, Indonesia
*Corresoponding author: putu.suwardika@yahoo.co.id
ABSTRACT
Creation of new superior varieties of mango through the empowerment of local genetic resources requires information about the potential properties of mango germplasm, including DNA polymorphism and its genetic diversity. This research aimed to obtain basic data on DNA polymorphism, genetic relationship, genetic similarity level, and molecular accession of Bali’s unique local mango. Sampling was conducted in four regencies in the Province of Bali. DNA preparation, PCR, and microsatellite analysis were carried out at the Laboratory of Genetics and Plant Breeding, Faculty of Agriculture, Gadjah Mada University. It found 44 accessions of mangoes in Bali. The test results with 10 pairs of SSR markers showed that all primers produced polymorphic loci. There were 825 amplified DNA bands. Primer of AY31 produced the highest number of loci, which were 14 loci, while AY21 produced the fewest loci, i.e., 3 loci. Forty-four mango accessions showed a genetic similarity coefficient of 0.27 to 0.97. At a coefficient of 0.27, accessions were divided into 2 major groups: group A and group B. Group A consisted of two accessions, namely, KRA-005 and BDG-006 (Mangifera foetida Lour.), which had a similar coefficient of 0.657. Group B was divided into two smaller groups, namely, groups B1 and B2, at a similarity coefficient of 0.342. Group B1 consisted of 39 accessions, while group B2 consisted of 3 accessions. Accessions Madu Anggur, Gading, Sambuk Mengwi, Kakul and Pakel Sulangai were identified as having unique alleles.
Keywords: Polimorphism, genetic similarity, Mango, DNA, Microsatellite
INTRODUCTION
Creation of new superior varieties (varietas unggul baru or VUB) of mango by empowering local genetic resources requires information about the potential properties of mango germplasm. Information about the potential properties is obtained through exploration and characterization of mango germplasm. Furthermore, the information obtained becomes the basis for consideration
in developing conservation, breeding, management and sustainable use of mango genetic resources.
Characterization of mango germplasm in Indonesia has been carried out by a number of researchers from Indonesian Tropical Fruit Research Institute and various universities in Indonesia, for example, characterizations that were conducted by Kostermans and Bompart (1993) in the West
Kalimantan and South Kalimantan regions, Sumiasri et al. (2006) in Madiun Regency, Rai et al. (2008) in the Bali region specifically for Mangifera Caesia Jack., Krisnawati (2008) in the Central Kalimantan region, Sumarsono et al. (2012) in Banyumas Regency, Fitawati et al. (2013) in the Central Sumatra region, and Oktavianto et al. (2015) in Kediri Regency. Characterization was also carried out on mango germplasm collection in the Mango Experimental Garden in Kampung Cukur Gondang, Pasuruan Regency (Maltuchah and Zainudin, 2007; Fitawati et al., 2010; Zainudin et al., 2010; Utami et al., 2012; Tasliah et al., 2013; Karsinah et al., 2015). However, given the vast territory of Indonesia and the limited budget and availability of research resources, not all mango germplasm can be characterized, including those mango germplasm in the Province of Bali. Bompard and Schnell (1997) reported that there were 5 species of mangoes found in the Province of Bali. The five species are Mangifera indica L., Mangifera foetida Lour., Mangifera caesia Jack, Mangifera odorata Griff., and Mangifera lalijiwa Kosterms. A quite indepth study of the characteristics of the Mangifera Caesia Jack was carried out by Rai et al. (2008). However, species of Mangifera indika L., Mangifera foetida Lour., Mangifera odorata Griff., and Mangifera lalijiwa Kosterms are still not
much studied so that basic data on DNA polymorphism, genetic similarity, genetic relationship, and the molecular uniqueness of mango germplasm in Bali are very limited.
This research aims to obtain basic data on DNA polymorphism, genetic relationship, and the level of genetic similarity, and to obtain accessions of unique local mangoes in Bali that molecularly and potentially may be developed as New Superior Varieties.
MATERIALS AND METHODS
Research Location and Time
Field research (on farm characterization) was conducted in 4 (four) regencies in the Province of Bali, Indonesia, namely, Buleleng, Karangasem, Badung, and Klungkung. Meanwhile, DNA preparation, PCR, and microsatellite analysis were conducted in the Laboratory of Genetics and Plant Breeding at the Faculty of Agriculture, Gadjah Mada University. The study was conducted for 13 months from July 2018 to August 2019.
Materials and Research Tools
Exploration equipments that were used included GPS, digital cameras, books of mango descriptors, scissors, label paper, plastic straps, paper bags, cool boxes, and stationery. Meanwhile, DNA analysis equipments were mortar, centrifugation
machines, micro pipettes, micro tubes, ovens, MJ Research PCT 100 PCR machines, gel electrophoresis, and Beckman Coulter® CEQ-8000 genetic analyzer. The materials used were young leaves, extraction buffer, EDTA, ice, lysis buffer, dithiothreitol, chloroform isoamil alcoholyl, sodium acetate, isopropanol, 70% ethanol, PCR buffer, forward primer, reverse primer, and Taq DNA Polymerase.
Determination of Sampling Location
Accession of Bali’s local mangoes was obtained through exploration at predetermined sampling locations. The sampling location was determined by the stratified random sampling method (Chaudhuri and Stenger, 2005). Sampling was done in two stages. The first stage was to determine the location of the district. Meanwhile, the second stage was to determine the sub-district sampling locations in each selected district. The research sample was taken from 4 (four) districts that were sorted by the number of sub-districts with local mango accessions. Each district was chosen from 4 sub-districts with the highest number of villages with mango accession. Mango plant leaf samples were taken from each accession that was found in each
village. Plants that were selected were plants that were more than 10 years old, in healthy conditions, and classified as the best among plants on the same accession in the vicinity.
Scope of Bali’s Local Mangoes
Determination of local mango accession refers to the scope proposed by Rai et al. (2016) that Bali’s local mangoes belong to the Bali’s local fruit group. As defined by Article 1 number 6 of the Bali Province Regional Regulation Number 3 of 2013, Bali’s local fruit referred to in this research refers to all types of fruits developed and cultivated in Bali.
Molecular Characterization
Young leaf samples of 3 pieces per accession were wrapped in aluminum foil, placed in a cool box filled with dry ice, then taken to the laboratory. Molecular characterization used 10 pairs of microsatellite primers, which had been tested on mango plants (Schnell et al., 2005; Tasliah et al., 2013). A list of sequences and types of microsatellite primer repeats are listed in Table 1.
Table 1. List of Sequences and Repetition Types of Microsatellite Primers Used in the Research
|
Primer |
Forward |
Reverse |
Repeats |
|
AY942818 |
CCACGAATATCAACTGCTGCC |
TCTGACACTGCTCTTCCACC |
(CT/AG)11 |
|
AY942821 |
TGTAGTCTCTGTTTGCTTC |
TTCTGTGTCGTCAAACTC |
(GTT/AAC)6 |
|
AY942825 |
CGAGGAAGAGGAAGATTATGAC |
CGAATACCATCCAGCAAAATAC |
(CGG/CCT)7 |
|
AY942827 |
GTTTTCATTCTCAAAATGTGTG |
CTTTCATGTTCATAGATGCAA |
(CT/AG)15 |
|
AY942828 |
CTCGCATTTCTCGCAGTC |
TCCCTCCATTTAACCCTCC |
(AG/CT)9 |
|
AY942829 |
GAACGAGAAATCGGGAAC |
GCAGCCATTGAATACAGAG |
(GTT/AAC)8 |
|
AY942831 |
TTTACCAAGCTAGGGTCA |
CACTCTTAAACTATTCAACCA |
(GA/TC)15 |
|
AJ635165 |
GATGAAACCAAAGAAGTCA |
CCAATAAGAACTCCAACC |
(TG)10 |
|
AJ635168 |
TTCTAAGGAGTTCTAAAATGC |
CTCAAGTCCAACATACAATAC |
(GT)9 |
|
AJ635176 |
TGCGTAAAGCTGTTGACTA |
GACAAGATAAACAACTGGAA |
(TG)11 |
Source: Scanell et al. (2005); Tasliah et al. (2013)
DNA Isolation and PCR Analysis
A total of 1 to 2 g of young leaves from each accession was extracted until DNA samples were obtained. The DNA isolation procedure used cetyl trimethyl ammonium bromide (CTAB) buffer extraction, following the Doyle and Doyle (1990) protocols. Dithiothreitol was added to prevent oxidation of phenolic compounds during extraction. The PCR reaction was carried out in a total volume of 20 μl.
DNA multiplication stages were initiation of denaturation at 95°C for 3 minutes, followed by 34 cycles consisting of denaturation at 94°C for 1 minute, primer attachment at 50°C for 1 minute, and extension of base at 72°C for 2 minutes (Schnell et al., 2005). The final stage of the PCR process was the final extension at 72°C for 5 minutes and incubation at 4°C. To see whether an amplified DNA appeared, electrophoresis using 2% agarose gel was performed. Primers, which produced bands in
agarose gel, were used for DNA fragment analysis.
Analysis of DNA Fragments
DNA fragments were detected on a genetic analyzer (Beckman Coulter® CEQ ™ 8000). The procedure for preparing PCR product samples for loading and procedures for running CEQ 8000 followed the protocol established by Thomson (2004). Samples were analyzed by multiplexing, i.e., mixing PCR amplification products with two kinds of green and blue fluorescent primers in one well. PCR products with a certain volume (according to the optimization results) of each primer were mixed in one well, then 0.5 μl of CEQ internal size standard (400 bp) labeled in red and SLS with a final volume of 40 μl were added.
Fragment size analysis results on CEQ 8000 were processed using CEQ Fragment Analysis software. Then, binning was carried out. Binning is grouping DNA
fragments (alleles) based on the number of repeats of certain DNA motifs (for example, repeat of threads from two, three, or four bases pairs) flanked by a pair of microsatellite primers.
Data Analysis
DNA concentration and purity of Bali's local mango accession were measured using the Gene Quant 1300 spectrophotometer. DNA concentrations obtained were high, i.e., more than 100 ng/µL, even most DNA oncentrations reached thousands of ng/µL. The purity of DNA compared to impurities, including protein impurity, was measured by the wavelength of λ260/280, while the purity of DNA compared to impurity in the form of RNA was measured by the wavelength of λ260/280. Good DNA purity ranges from 1.8 to 2.2.
Analysis of mango germplasm accessions was based on the level of allele similarity between accessions, using the genetic distance formula described by Nei et al. (1983). The dendrogram was generated from the binary data of the mango plant molecular scoring results using the unweighted pair group method with arithmetic mean (UPGMA) using NTSYS 2.02 software.
RESULTS AND DISCUSSION
Findings of Bali’s Local Mango Accession
Exploration results found 44 accessions of Bali’s local mangoes, with the following details: 29 accessions were found in Buleleng Regency, 6 accessions were found in Badung Regency, 5 accessions were found in Karangasem Regency, and 4 accessions were found in Klungkung Regency (Table 2). Thirty four of them, or 77.27%, were landrace accessions. The rest were non-landrace accessions. Landrace accessions include Amplemsari (Legong), Bila, Bikul, Gedang Bontihing, Santog, Manggis, Dodol, Depeha, Sanih, Kunyit, Poh Wani Kubutambahan, Gandarasa, Amplemsari Tejakula, Gula, Kunahan, Amplem Taluh, Pudak, Ijo Bukit, Gading, Sambuk Mengwi, Kakul, Gedang Kelan, Lembat Tianyar, Taluh Tianyar, Siap, Santen Banjarangkan, Lembongan, Pelem Nusa Penida, Sakti, Eni Bontihing, Eni Ban, Pakel Sibetan, and Pakel Sulangai. Meanwhile, those that were included in non-landrace accessions were Gedong Gincu, Brazil, Parkit, Shailon, Arumanis 143, Apel, Manalagi, Golek, Madu Anggur, and Lalijiwa.
Table 2. Accessions of Bali’s Local Mangoes
|
No |
Field ID |
Local Name |
Origin |
No |
Field ID |
Local Name |
Origin |
|
1 |
BLL-001 |
Amplemsari (Legong) |
Buleleng |
23 |
BLL-023 |
Gula Bengkala |
Buleleng |
|
2 |
BLL-002 |
Gedong Gincu |
Buleleng |
24 |
BLL-024 |
Kunahan |
Buleleng |
|
3 |
BLL-003 |
Brazil |
Buleleng |
25 |
BLL-025 |
Amplem Taluh |
Buleleng |
|
4 |
BLL-004 |
Parkit |
Buleleng |
26 |
BLL-026 |
Pudak |
Buleleng |
|
5 |
BLL-005 |
Shailon |
Buleleng |
27 |
BDG-001 |
Ijo Bukit |
Badung |
|
6 |
BLL-006 |
Arumanis 143 |
Buleleng |
28 |
BDG-002 |
Gading |
Badung |
|
7 |
BLL-007 |
Bila |
Buleleng |
29 |
BDG-003 |
Sambuk Mengwi |
Badung |
|
8 |
BLL-008 |
Bikul |
Buleleng |
30 |
BDG-004 |
Kakul |
Badung |
|
9 |
BLL-009 |
Apel |
Buleleng |
31 |
BDG-005 |
Gedang Kelan |
Badung |
|
10 |
BLL-010 |
Manalagi |
Buleleng |
32 |
KRA-001 |
Lembat Tianyar |
Karangasem |
|
11 |
BLL-011 |
Gedang Bontihing |
Buleleng |
33 |
KRA-002 |
Taluh Tianyar |
Karangasem |
|
12 |
BLL-012 |
Santog |
Buleleng |
34 |
KRA-003 |
Siap |
Karangasem |
|
13 |
BLL-013 |
Manggis |
Buleleng |
35 |
KLK-001 |
Santen Banjarangkan |
Klungkung |
|
14 |
BLL-014 |
Golek |
Buleleng |
36 |
KLK-002 |
Lembongan (Nusa) |
Klungkung |
|
15 |
BLL-015 |
Dodol |
Buleleng |
37 |
KLK-003 |
Pelem Nusapenida |
Klungkung |
|
16 |
BLL-016 |
Depeha |
Buleleng |
38 |
KLK-004 |
Sakti Nusapenida |
Klungkung |
|
17 |
BLL-017 |
Sanih |
Buleleng |
39 |
BLL-027 |
Eni (Kweni) Bontihing |
Buleleng |
|
18 |
BLL-018 |
Kunyit |
Buleleng |
40 |
KRA-004 |
Eni (Kweni) Ban |
Karangasem |
|
19 |
BLL-019 |
Putih |
Buleleng |
41 |
KRA-005 |
Pakel Sibetan |
Karangasem |
|
20 |
BLL-020 |
Wani Kubutambahan |
Buleleng |
42 |
BDG-006 |
Pakel Sulangai |
Badung |
|
21 |
BLL-021 |
Ganda Rasa |
Buleleng |
43 |
BLL-028 |
Madu Anggur |
Buleleng |
|
22 |
BLL-022 |
Amplemsari Tejakula |
Buleleng |
44 |
BLL-029 |
Lalijiwa |
Buleleng |
Based on species, 44 accessions of Bali’s local mangoes come from 4 species, namely, Mangifera indica L., Mangifera odorata Griff., Mangifera foetida Lour., and Mangifera lalijiwa Kosterms. Accession of the species of Mangifera indica L. dominates mango germplasm in Bali with 38 accessions or 86.36%. This finding corroborates the results of research by Bompard and Schnell (1997) that reported that there were 5 species of mangoes found in Bali. The five species were Mangifera indica L., Mangifera foetida Lour., Mangifera caesia Jack, Mangifera odorata Griff., and Mangifera lalijiwa Kosterms. Specifically on Mangifera caesia Jack, Rai et al. (2008) have conducted quite in-depth studies. However, on the other 4
species, no in-depth molecular studies have been carried out.
DNA Concentration and Purity
DNA concentration and purity of 44 Bali’s local mango accessions were measured using the GeneQuant 1300
spectrophotometer. DNA concentrations obtained were high, i.e. more than 100 ng/µL. Most of concentrations even reached thousands of ng/µL. Good DNA purity ranges from 1.8 to 2.2. From the results of the quantification, it was known that the DNA purity of the 44 accessions of the mango analyzed was quite good because it was in the range 1.8 - 2.2.
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 7 No. 1 • September 2019
DNA Polymorphism
Test results of 44 Bali’s local mango accessions with ten pairs of SSR markers indicated that all primers that were used produced polymorphic loci. There were 825 DNA bands that were successfully amplified. The primer that produced the most loci was AY31, which produced as many as 14 loci, while the primer that produced the least locus was AY21, which produced as many as 3 loci only.
Genetic Relationship and Similarity Level
The dendrogram was generated from the binary data of the mango accession molecular scoring results, as presented in Fig. 1. From the dendrogram, it was known that the genetic similarity coefficient value of 44 accessions of Bali’s local mango ranged from 0.27 to 0.97. The greater the similarity coefficient value is, the more similar DNA band patterns between accessions become,
eISSN: 2655-9994 pISSN: 2303-3371 https://doi.org/10.24843/IJBB.2019.v07.i01.p05 and that means the accessions are increasingly similar. In this dendrogram, it can be seen that 44 mango accessions were divided into two large groups at a coefficient of 0.27, namely, group A (2 accessions) and group B (42 accessions). Group A consisted of two accessions, namely, KRA-005 and BDG-006 (species of Mangifera foetida Lour.), which had a similarity coefficient of 0.657. Group B was divided into two smaller groups, namely, groups B1 and B2 at a similarity coefficient of 0.342. Group B1 consisted of 39 accessions, while group B2 consisted of 3 accessions (BDG-002, BDG-003, and BDG-004). Among these 44 accessions, there were two very similar accessions, namely, BLL-006 and BLL-016, which were located in the similarity coefficient of 0.97.
BLL-O01 BLL-013 BLL-OIO 8LL-012
BLL-014 BLL-003 8LL-0O5
BLL-009 BLL-0O4 SLL-OOS
BLL-O07 BLL-002
KLK-002 BLL-O11 BLL-OOS BLL-OIG
BLL-OI7 BLL-OI5
BLL-026 BLL-020
BLL-OI8 BLL-021 BLL-∪23
BLL-022 BLL-013
BLL-024 BLL-025
BDG-005 KRA-001 KRA-002 KRA-003 BDG-001
KLK-O01 KLK-003
KLK-004 BLL-029 BLL-028
BLL-027 KRA-004 BDG-002 BDG-003
BDG-004 KRA-005 RDG-OOS
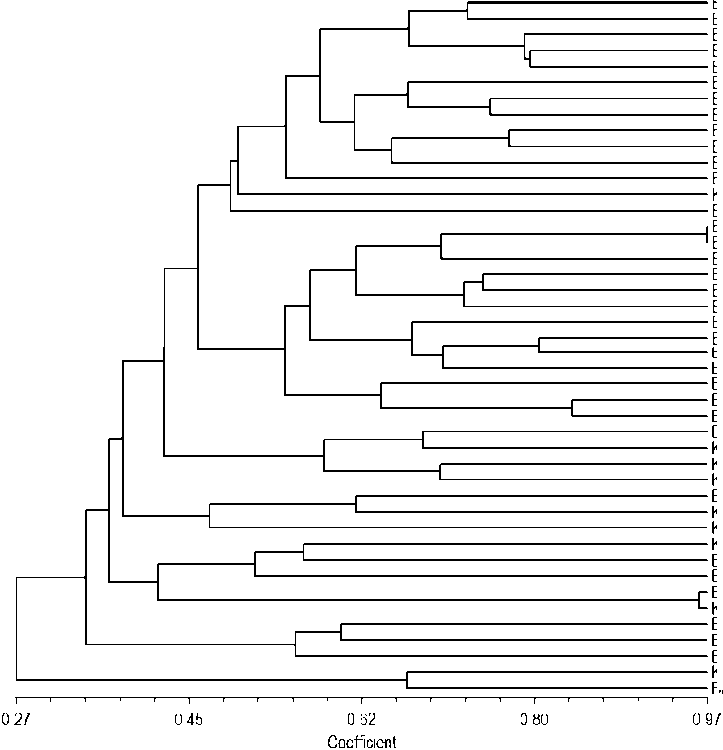
Fig. 1. Dendrogram of 44 Bali’s local mango accessions BLL, BDG, KRA, and KLK are accessions code
The principal coordinates analysis was used to find out the tendency of grouping of 44 Bali’s local mango accessions based on their species. Forty-four accessions of Bali’s local mangoes belong to the species of Mangifera indica L. (marked by a blue dot), Mangifera foetida Lour. (marked by an
orange dot), Mangifera odorata Griff. (marked by a gray dot), and Mangifera lalijiwa Kosterms (marked by a yellow dot), as can be seen in Fig. 2. The tendency of this grouping is generated from the value of Genetic Distance (GD) using GenAlEx software 6.503.
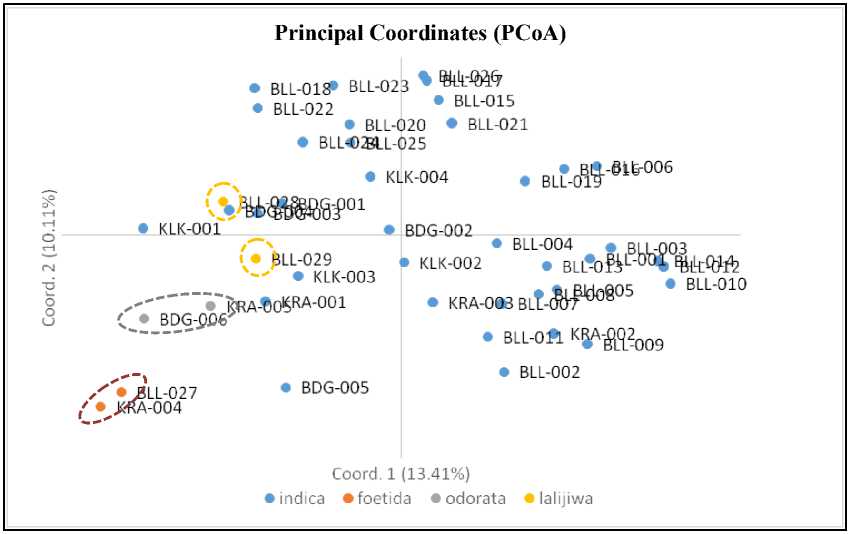
Fig. 2. Principal coordinates analysis
of 44 Bali’s local mango accessions with 10 SSR markers BLL, BDG, KRA, and KLK are accessions code
Fig. 2 shows that mango accessions generally tended to spread mainly to Mangifera indica L. species. However, Mangifera foetida Lour. species (marked by an orange dot) and Mangifera odorata Griff. (marked by a gray dot) are located close together according to their respective species groups. The Mangifera lalijiwa Kosterms species are located close together, but are among the indica accession groups.
The Unique Accession of Mango Germplasm
Among 44 accessions of Bali’s local mangoes, there are five individuals that have one unique allele, as presented in Table 3.
The five accessions are BLL-028, BDG-002, BDG-003, BDG-004, and BDG-006. These alleles were mostly amplified by different primers, but some were identified in the same primers at different loci (AJ68 in accessions BDG-004 and BDG-003). Unique alleles are alleles that are only owned by one individual/accession at a specific locus size, and are not owned by another individual. BLL-028 (Madu Anggur) is a superior variety that has been released by the government or is classified as non-landraces. Whereas, BDG-002 (Gading), BDG-003 (Sambuk Mengwi), BDG-004 (Kakul), and BDG-006 (Pakel Sulangai) are accessions of landraces. In principle, accessions that have
unique alleles can be a characteristic of the accessions concerned and have the potential to be developed as plant breeding material
and combined with other superior properties so that in the long run, they can be developed into New Superior Varieties.
Table 3. List of Mango Accessions that have Unique Alleles
|
Accession Code (Field ID) |
Primer and Locus Size (bp) |
|
BDG-002 |
AY27 (255 bp) |
|
BLL-028 |
AY29 (450 bp) |
|
BDG-006 |
AY31 (270 bp) |
|
BDG-004 |
AJ68 (260 bp) |
|
BDG-003 |
AJ68 (310 bp) |
The findings of this study can facilitate improved management of local Bali mango germplasm conservation and help find strategies for future breeding activities. The same opinion was expressed by Bora et. al. (2018) based on the results of his study of 19 mango genotypes in India, Razak et. al. (2019) from the results of his research on mango germplasm in Malaysia, Riaz et. al. (2018) based on the results of mango research in Pakistan, and Yamanaka et. al. (2019) based on the results of his study of 120 mango genetic resources in Japan.
CONCLUSION
It can be concluded from this study that 44 mango accessions were found in Bali. Thirty four accessions or 77.27% are landrace accessions. The rest are nonlandrace accessions. The accession of Bali’s local mangoes originated from 4 species, namely, Mangifera indica L., Mangifera odorata Griff., Mangifera foetida Lour., and Mangifera lalijiwa Kosterms. Ten primers
produced polymorphic loci. There were 825 amplified DNA bands. AY31 Primer produced the highest number of loci, which was 14 loci, while AY21 Primer produced the fewest loci, i.e., as many as 3 loci. Forty-four mango accessions showed a genetic similarity coefficient of 0.27 to 0.97. At a coefficient of 0.27, it was divided into 2 major groups, namely, group A and group B. Group A consisted of two accessions, namely, KRA-005 and BDG-006 (species of Mangifera foetida Lour.), which had a similarity coefficient of 0.657. Group B was divided into two smaller groups, namely, groups B1 and B2 at a similarity coefficient of 0.342. Group B1 consisted of 39 accessions, while group B2 consisted of 3 accessions. Accessions Madu Anggur, Gading, Sambuk Mengwi, Kakul and Pakel Sulangai were identified as having unique alleles.
ACKNOWLEDGMENTS
We would like to thank the Director of Research and Community Service in the Directorate General of Research and Development Strengthening, Ministry of Research, Technology and Higher Education for the Doctoral Dissertation Research Program that has been given, in accordance with the 2018 Budget Year Research Contract Number: 0705/K8/KM/2018.
REFERENCES
Bompard, J. M., & Schnell, R. J. (1997). Taxonomy and systematics. In: Litz RE (Ed.) The mango: botany,
production and uses. CAB International, Wallingford. pp. 21-47.
Bora, L., Singh, A. K., Kumar, A., &
Metwal, M. (2018). Morphological and microsatellite marker based polymorphic assesment of genetic diversity and reationship of mango (Mangifera indica L.). Indian Journal of Biotechlogy, 17, 91-100.
Chaudhuri, A., & Stenger, H. (2005). Survey Sampling Theory and Methods.
Second Edition. Chapman dan
Hall/CRC. Taylor dan Francis Group. 371 p.
Doyle, J. J., & Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus 12, 13-15.
Fitawati, Suwita, A., Sofiyanti, N.,&
Herman. (2013). Eksplorasi dan karakterisasi keanekaragaman plasma nutfah Mangga (Mangifera)di
Sumatera Tengah. Prosiding
Semirata FMIPA Universitas Lampung, 2013: Hal 307-311(In
Indonesian).
Karsinah, Sutanto, A., Mansyah, E., Rebin, Wahyuni, D., & Tasliah. (2015).
Identifikasi marka molekuler untuk
seleksi genotipa Mangga tahan rontok dan ukuran buah besar. Laporan Hasil Penelitian Balitbu Tropika, Solok (In Indonesian).
Kostermans, A. J. G. H., & Bompart, J.M. (1993). The mangoes their botany, nomenclature and utilization.
International Board for Plant Genetic (IBPGR). Academic Press.
Maltuchah & Zainudin, A. (2007). Variasi genetik beberapa kultivar mangga dengan menggunakan penanda molekuler Random Amplified Polymorphic DNA. Prosiding
Seminar Nasional Hortikultura, Surakarta, 17 Nopember 2007 (In Indonesian).
Nei, M., Tajima, F., & Tateno, Y.
(1983). Accuracy of estimated
phylogenetic trees from molecular
data. II. Gene frequency data. J. Mol. Evol. 19(2), 53-70.
Oktavianto, Y., Sunaryo, & Suryanto, A. (2015). Karakterisasi tanaman
mangga (Mangifera Indica L.)
Cantek, Ireng, Empok, Jempol di Desa Tiron, Kecamatan Banyakan, Kabupaten Kediri. Jurnal Produksi Tanaman, 3(2), 91-97 (In
Indonesian).
Rai, I.N., Wijana, G., & Semarajaya, C. G. A. (2008). Identifikasi variabilitas genetik wani Bali (Mangifera caesia Jack.) dengan analisis penanda RAPD. Jurnal Hortikultura, 18(2), 125-134 (In Indonesian).
Rai, I.N., Wijana, G., Sudana, I P., Wiraatmaja, I W., & Semarajaya, C. G. A. (2016). Buah-buahan lokal Bali. Jenis, pemanfaatan dan potensi pengembangannya. Palawasari,
Denpasar. 280 hal (In Indonesian).
Razak, S. A. B., Azman, N. H. E. N., Ismail, S. N., Yusof, M. F. M., Arrifin, M. A. T., Sabdin, Z. H. M., Hassan, M. H. M., Nasir, K. H., Sani, M. A., & Abdullah, N. (2019). Assessment of diversity and population structure of mango (Mangifera indica L.)
germplasm based on microsatellite (SSR) markers. Australian Journal of Crop Science, 13(02), 315-320.
Riaz, R., A.S. Khan, K. Ziaf, and H.M.N. Cheema. (2018). Genetic diversity of wild and cultivated mango Genotypes of Pakistan using SSR markers. Pakistan Jounal Agricultural Science, 55(1), 83-93.
Schnell, R.J., Olano, C. T., Quintanilla, W. E., & Meerow, A. W. (2005).
Isolation and characterization of 15 microsatellite loci from mango (Mangifera indica L.) and crossspecies amplification in closely related taxa. Mol. Ecol. Notes 5, 625627.
Sumarsono, Suparjana, T. B., & Purwati, E. S. (2012). Karakterisasi morfologi dan pola pita izosim varietas mangga (Mangifera sp.) di Kabupaten
Banyumas. Prosiding Seminar Nasional Pengembangan Sumber Daya Pedesaan dan Kearifan Lokal Berkelanjutan II, Purwokerto 27-28 Nopember 2012 (In Indonesian).
Sumiasri, N., Rijadi, J., & Priadi, D. (2006). Variasi jenis dan kultivar mangga di Madiun dan sekitarnya;
pengembangan dan permasalahannya. Biodiversitas, 7(1), 39-43 (In
Indonesian).
Tasliah, Rijzaani, H., Hariyadi, T. Z. P., Rebin, Ma’sumah, & Silitonga, T. S. (2013). Analisis keragaman genetik 161 aksesi mangga Indonesia menggunakan marka mikrosatelit. Jurnal Agro Biogen, 9(3), 125-134 (In Indonesian).
Thomson, M.J. (2004). Microsatellite fragment sizing on the CEQ 8000: BB Biogen standard operating procedure series. Indonesian Center for Agricultural Biotechnology and Genetic Resources Research and Development. Bogor. Indonesia. p. 110.
Utami, D.W., Santoso, T. J., & Hidayatuni, N. (2012). Sidik jari DNA plasma nutfah mangga berdasarkan analisis fragmen marka SSR (Simple Sequence Repeat) berlabel. Jurnal Hortikultura Indonesia, 3(1), 49-57 (In Indonesian).
Yamanaka, S., Hosaka, F., Matsumura, M., Makishi, Y. O., Nashima, K., Urasaki, N., Ogata, T., Shoda, M., & Yamamoto, T. (2019). Genetic diversity and relatedness of mango cultivars assessed by SSR markers. Breeding Science, 69, 332–344.
Zainudin, A., Maftuchah, Martasari, C., & Santoso, T. J. (2010). Keragaman genetik beberapa kultivar tanaman mangga berdasarkan penanda
molekul mikrosatelit. Makalah
Lengkap Kongres Ketiga Komisi Daerah Sumber Daya Genetik. Hotel Singgasana, Surabaya, tanggal 3‐5 Agustus 2010 (In Indonesian).
56 • FACULTY OF AGRICULTURE, UDAYANA UNIVERSITY
Discussion and feedback