MORPHOLOGY, PHYSIOLOGY AND MOLECULAR CHARACTERISTICS OF OIL PALM (Elaeis guineensis Jacq.) ENDOPHYTIC Bacillus sp.
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 5 No. 1 • September 2017
ISSN: 2303-3371
https://doi.org/10.24843/IJBB.2017.v05.i01.p07
MORPHOLOGY, PHYSIOLOGY AND MOLECULAR CHARACTERISTICS OF OIL PALM (Elaeis guineensis Jacq.) ENDOPHYTIC Bacillus sp.
Fifi Puspita1,2*, Hadiwiyono1, Susilo Hambeg Poromorto 1, and
Dewi Indriyani Roslim3
-
1Department of Agriculture Science, Graduated School of Sebelas Maret University, Surakarta, Indonesia
-
2Department of Agrotechnology, Faculty of Agriculture, Universitas Riau, Pekanbaru, Indonesia
-
3Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Riau, Pekanbaru, Indonesia
*Corresponding authors: fipspt@gmail.com
ABSTRACT
Endophytic bacteria are the bacteria that live in plant tissues. In oil palm tissue there are many types of endophytic bacteria and have a role that can be beneficial for the plant, one of them is endophytic Bacillus sp. The aim of these research was to obtain morphology, physiology and molecular characteristics of endophytic Bacillus sp. originating from oil palm tissue. Sampling was done by random simple sampling method. Isolation of bacteria was performed on plant tissues such as roots, midribs, stems and leaves of oil palm plants. The results of morphological characterization such as colony color, colony shape and colony edge show similarity in each isolate but there are differences in the surface morphology of the colony, where there are 6 isolates with convex surface and 6 isolates with flat shape. Physiological test results such as catalase test, oxidase test, starch hydrolysis test, motility test and temperature effect test on bacterial growth showed positive results in each isolate. Molecular characterization using 16S rRNA primers based on BLASTn shows that all isolates tested have similarities with Bacillus sp. Based on the phylogenetic tree it was found that the endophytic bacteria of Ba-B2 isolates were associated with Bacillus flexus with 100% consistency index grouped at a distance of 0.03 and Ba-P2 isolates were related to Bacillus substilis at a distance of 0.01 with an 89% consistency index.
Keywords: 16S rRNA, Bacillus sp., Blastn, endophytic bacteria, oil palm
INTRODUCTION
The oil palm plant (Elaeis guineensis Jacq.) in Indonesia is widely cultivated. However, the production of oil palm crops in Indonesia is always attributed to fluctuating yield, due to
biotic factor. Basal stem rot disease that caused by Ganoderma boninense Pat. is the important one.
Control over the years done by using chemical fungicides does not show satisfactory results. Application of
biological fungicides has also been done by applying Bacillus sp. origin of the rhizosphere but also has not addressed unsatisfactory results. Alternatives to overcome the spread of basal stem rot disease can be done by isolating endophytic bacteria from palm oil plant tissue.
Endophytic bacteria will colonize so as to inhibit the growth of pathogenic microbes through the mechanism of space competition and nutrition (Pal et al., 2012). Endophytic bacteria can also act as biological fertilizers. Endophytic bacteria have several other roles such as N2 inhibitors from the air, producing phytohormones such as Indole-3 Acetic Acids (IAA) and sitokinin that can spur growth (Setiawati et al., 2009).
Endophytic bacteria that can be isolated from plant tissue organs are Bacillus sp. Isolation of Bacillus sp. Endophytes derived from oil palm crops can be done from the root, stem, midrib and leaves (Tarabilly et al., 2003).
MATERIALS AND METHODS
The study was conducted from February 2016 until May 2016 in the Business Unit Biofertilizer and biopesticides Genetics Laboratory, Faculty of Agriculture and Department of Biology, State University of Riau Pekanbaru Simpang Jalan Baru Binawidya km 12.5 Panam, District Charming Pekanbaru. In this experiment, 12 isolates of endophytic bacteria that 3 isolates of root tissue (A1, A2, A3), 3 isolates of stem tissue (B1, B2, B3), 3 isolates of leaf tissue (D1, D2, D3), and 3 Isolates from the lymph tissue (P1, P2, P3).
Morphological characterization was done by observing bacterial colonies grown on Nutrient Agar medium. Morphological characterization of endophytic bacteria includes colony color, form colonies, colonies edge, the surface of the colonies, Gram bacteria and bacterial endospores), Characterization physiologically covering of the test gram, catalase, oxidase, the need for oxygen
(oxidative-fermentation), the hydrolysis of gelatin, starch, formation of levan, test Voges Proskauer, arginine dehidrolase, motility, tolerance bacterial growth at some temperature, pH and concentration of HCl, the use of and overhaul of carbon compounds, citric and nitrogen (Lelliot and Stead, 1987).
Molecular characterization begins by executing bacterial DNA extraction using a bacterial DNA isolation kit (Geneaid DNeasy Blood & Tissue). The obtained DNA was then amplified by PCR technique using BSF820_F (forward) 5'-AGA GTT TGA TGG CTC AG-3 TGA pair and BSR1521_R (reverse) 5'-AAG GAG GTG ATC CAG CCG CA-3 '. The PCR reagent used for each reaction was 5 μL PCR buffer, 2.5 μL mM dNTP, 1 μL primary forward, 1 μL reverse primer, 0.5 μL taq polymerase, 39 μL dH2O and 1 μL endophytic bacterial DNA. Further amplification was done on PCR machine (thermo cycler) with denaturation program at 94 ° C for 30 seconds, annealing at 52 °
C for 45 seconds, elongation at 72 ° C 1 minute 30 seconds, and post PCR at 72 ° C for 10 minute. The PCR product was identified by electrophoresis using agarose gel.
Measurement of DNA fragments using a 1kb DNA ladder marker. The PCR product is then sequenced. The results of the sequence of nucleotide sequences of 16S ribosomal DNA were analyzed by BLAST (Basic Local Alignment Search Tool) software found on the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov) site to find out the closest familial level to the bacteria present in In the GenBank database (NCBI). Further sequence of 16S ribosomal DNA from bacteria was taken for the analysis of phylogenetic trees. Multiple Sequence Alignment and phylogenetic tree construction were analyzed using the MEGA6 program. The phylogenetic tree was prepared by the neighbor-joining algorithm method, with stability grouping using bootstrap analysis
with 1000 repetitions. The data obtained are analyzed descriptively and taken in the form of graphs and drawings.
RESULTS AND DISCUSSION
All endophytic bacterial isolates have uniform colors and shapes, which are white and round in shape. Further morphological characteristics are presented in Table 1. This characterization of 12 isolates was derived from the same genus of endophytic bacteria, the genus Bacillus sp. According to Hatmati (2000), Bacillus spp. Has the edges of the colony of various kinds and uneven, the surface is rough and not slimy, there is even tend to dry and powder, colonies large and not shiny.
The identification result of 12 isolates of Bacillus sp. The endophytes observed by characterizing morphology and physiology resemble those characteristic of the Bacillus subtillis species. These results include positive
endospora, Gram positive bacteria, positive catalase test, positive oxidation test, positive starch hydrolysis test, positive motility test and 37ºC optimum temperature. The results of all the tests indicate the same characterization with positive results. The physiological and biochemical properties of the six isolates of Bacillus spp. Are presented in Table 2.
In the oxidase test conducted on 12 isolates of Bacillus sp. Endophytes showed positive results in all isolates. From this oxidase test, Bacillus sp endophytic bacteria can produce oxidase enzyme. Enzyme oxidase in Bacillus sp. Endophytes play an important role in transport electrons during aerobic respiration, cytochrome by oxygen molecules. The enzyme oxidase produced by aerobic facultative aerobic bacteria and microaerophilic in Bacillus sp. Causing these bacteria to be able to utilize the available carbon source (Priyani, 2006).
Table 1. Characteristics of bacterial morphology Bacillus sp. Endophytes isolated from oil palm trees on NA media.
|
Isolate codes |
Morphological Characteristics |
|
Gram Endospora Surface Shape Color | |
|
A1 A2 |
+ + flat Round White + + Convex Round White |
|
A3 B1 B2 |
+ + flat Round White + + Convex Round White + + Convex Round White |
|
B3 D1 D2 |
+ + Convex Round White + + flat Round White + + flat Round White |
|
D3 P1 P2 |
+ + Convex Round White + + Convex Round White + + flat Round White |
|
P3 |
+ + flat Round White |
In the 12 isolates tested positive results in all bacterial isolates of Bacillus sp. Endophytes. These results are seen from the bright zones that appear after iodine salt is added. These results address Bacillus sp. Capable of hydrolyzing starch and having an amylase enzyme in which these results are seen from bright zones formed on bacterial colonies that have been added iodine salts. Bright zone can be seen clearly with the addition of indicator of iodine salt, starch that does not hydrolyze the amylase enzyme will be
blackish black with the addition of Gram iodine. This is because the triodide ions of the iodine gram solution act with a straight chain amylose helix forming a color complex. The color produced by the iodine reaction with the polysaccharide depends on the three-dimensional structure of the polysaccharide. While the starch that has been hydrolyzed by the amylase enzyme will form a bright zone because glucose cannot act with the colored complex triodide ion complex (Koolman and Roehm, 2005).
Table 2. Physiological characteristics of endophytic bacteria
|
Parameters |
Endophitic bacterial isolates | |||||||||||
|
A1 |
A2 |
A3 |
B1 |
B2 |
B3 |
D1 |
D2 |
D3 |
P1 |
P2 |
P3 | |
|
Physiological Test Catalase Test |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Oxidation Test |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Hydrolysis of |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
starch Growth at 35 ° C |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Voges test |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Arginine |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Dehydrolase Motility Test |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Bacillus sp. Has motile properties and is also inmotile. Bacillus sp. Motile species are Bacillus subtilis and Bacillus thurgenensis. Bacillus subtilis is said to be motile because this bacterium can move with a set of flagella attached to both poles. Motile properties caused by the existence of a motor tool whip called flagell so that bacteria cells can swim in the water environment. Motility of most
types of motile bacteria at relatively low temperatures is 15-25 º C and may not be motile at 37 ºC. Some bacteria can perform very smooth slides that occur only when contact with solids (Tarigan, 1988).
The electrophoresis results from the extraction sample and endophytic bacteria showed the total obtained DNA that was marked with a thick band above 10000
Bp. Total DNA from some isolates showed that there were also DNA smears or degradation, but the DNA could still be used as a template on PCR (Fig. 1A).
Result of amplification visualization of whole isolate of Bacillus spp. used on the basis of the 16S rRNA gene showed DNA bands parallel to the size of ± 1500 bp (Fig. 2B). Visualization was performed using a 1.5% agarose gel and observed under a UV transiluminator. Identification followed by sequencing. The sequencing is based on the sequence of the 16S rRNA gene which then the DNA base sequence is required to identify the isolates identified by the existing database in GeneBank NCBI using the BLAST program.
Based on the results of BLAST, samples D1, D2, P1, P2, and P3 have similarities with Bacillus subtillis with different identical values in each isolate. Isolate P2 has an ident value of 98% with
accession code HQ123475.1. Isolate B2 has similarities with Bacillus flexus with accessory code KP866883.1 with a 94% ident value. The B3 sample has a resemblance to the uncultured Bacillus sp. With an access code GQ890463.1 with an ident value of 88%. While other isolates have similarities with Bacillus sp. With various identical values (Table 3).
On the phylogenetic tree (Fig. 2) it can be seen that in the first cladrogram (I) it consists of 12 isolates tested (A1, A2, A3, B1, B2, B3, D1, D2, D3, P1, P2, P3), Bacillus Flexus (GenBank), Bacillus subtilis (GenBank) and Bacillus sp. (GenBank). The second cladrogram (II) contains only Escherichia coli (GenBank) because E. coli is only used as a test strain. Seki et al. (1978) in his journal also used Escherichia coli as a comparative strain in the taxonomy of the Bacillus genus.
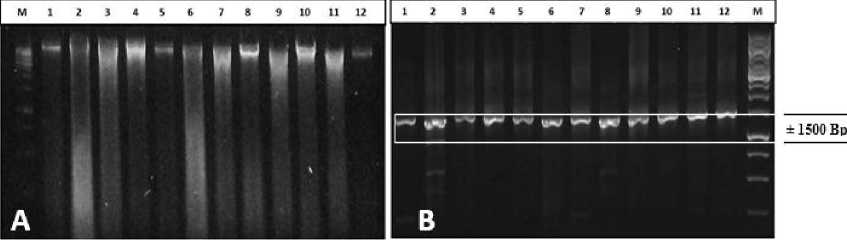
Fig. 1. Total DNA (A) of 12 samples of bacterial endophytic bacteria isolate in electrophoresis at 1.2% agarose gel. Information : (M) 1kb DNA ladder, (1) A1, (2) A2, (3) A3, (4) B1, (5) B2, (6) B3, (7) D1, (8) D2, (9) D3, (10) P1, (11) P2, (12) P3
In the first clan program is divided into two subkladrograms (Ia) and (Ib). The first subgrogram (Ia) consists of isolates B2, B1, A3, A1, P1, A2, D2, Bacillus flexus (GenBank), Bacillus sp. (GenBank). Bacillus flexus (GenBank) with isolate B2 grouped at a distance value of 0.02 with 100% bootstrap consistency value. Isolate B1 is also clumped with Bacillus sp. (GenBank) at a distance value of 0.01 with a 100% bootstrap consistency value. Isolate A2 grouped with isolate D2 at a distance value of 0.03 with a bootstrap 99%
consistency value. Based on morphological and physiological characteristics, all endophytic bacterial isolates belong to the genus Bacillus. This is reinforced by the molecular Blast results showing kinship with Bacillus sp seen at high identical values. According to Drancourt (2000) based on the 16S rRNA gene sequence data, if the 99% ident value can be said that the comparable species are the same species, whereas if the 97% ident value can be stated that the comparable isolate belongs to the same genus.
Table 3. Results of DNA sequence alignment of the 16S rRNA gene in all samples of endophytic bacteria of oil palm
|
Isolat |
Description |
Accession |
Max Score |
Total Score |
Query Cover |
E | |
|
Value |
Ident | ||||||
|
A1 |
2111 |
2111 |
84% |
0.0 |
95% | ||
|
A2 |
KJ948345.1 |
1465 |
1465 |
77% |
0.0 |
87% | |
|
A3 |
1860 |
1860 |
87% |
0.0 |
92% | ||
|
B1 |
2489 |
2489 |
93% |
0.0 |
99% | ||
|
B2 |
Bacillus flexus strain ASR-9 16S ribosomal RNA gene, partial |
2093 |
2093 |
94% |
0.0 |
94% | |
|
B3 |
1541 |
1541 |
79% |
0.0 |
88% | ||
|
D1 |
1243 |
1243 |
77% |
0.0 |
84% | ||
|
D2 |
1456 |
1456 |
65% |
0.0 |
88% | ||
|
D3 |
LC133718.1 |
1344 |
1344 |
99% |
0.0 |
82% | |
|
P1 |
1525 |
1525 |
67% |
0.0 |
88% | ||
|
P2 |
2360 |
2360 |
87% |
0.0 |
98% | ||
|
P3 |
1182 |
1182 |
95% |
0.0 |
80% | ||
In the second subkladrogram (Ib) consists of isolates P3, D3, D1, B3, P2, Bacillus subtilis (GenBank). Isolate P2 grouped with Bacillus subtilis (GenBank) at a distance value of 0.02 with a 100% bootstrap consistency value. Other isolates form different branches with different consistency values and at different distance values as well. The higher the value of the ident it can be said that the
higher the similarity of the aligned sequence. The Query cover value is the percentage of the corresponding nucleotide sequence length compared to the database contained in BLASTn. Ident is the highest value of the identity percentage or the match between the query sequence and the aligned database sequence (Miller, 1990).
BaciNus sp. 2BSG-R2A-9 ∣ AB5337β1.1
BaciNus SUbtNis strain RS8102 ∣ HQ123475.1
BaciNus Nexus strain ASR-9 ∣ KP8M883.1 B2
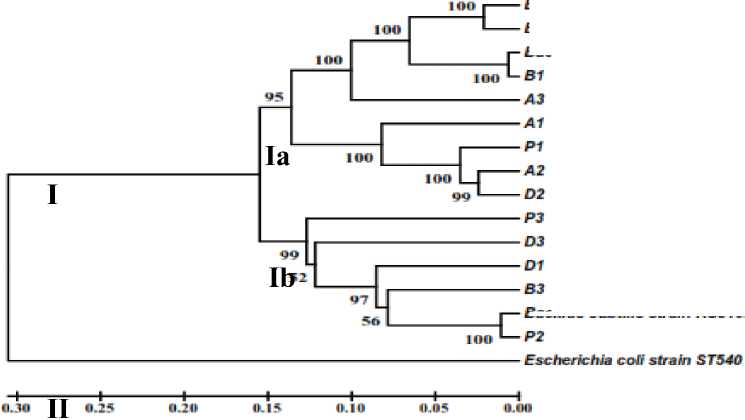
Fig. 2. Dendrogram between bacterial isolates with several species of Bacillus (GenBank) and Escherichia coli (GenBank) as a comparison based on distance matrix with UPGMA method with bootstrap 1000 times
Discussions
It can be concluded that morphological characterization shows 12 isolates of endophytic bacteria having the same color and shape (white and round), gram + bacteria, having endospores, with different surfaces, ie convex (A2, B1, B2, B3, D3, P1) and Flat (A1, A3, D1, D2, P2, P3). 12 isolates showed the same physiological characteristics test, i.e. positive catalase, positive oxidation, positive starch hydrolysis, positive motility and optimum temperature of 37ºC. Molecular characterization shows the entire isolate of endophytic bacteria are Bacillus genus. Endophytic bacteria isolate B2 is closely related with Bacillus flexus and P2 isolate is closely related to Bacillus substilis.
REFERENCES
Drancourt, M., Bollet, C., Carlioz, A., Martelin, R., Gayral, J. P., & Raoult, D. (2000). 16S ribosomal DNA
sequence analysisi of large collection of environmental and clinical unidentifiable bacterial
isolates. J Clin Microbioli, 38 :
3623-3630.
EI-Tarabilyl, K. A., Nassar, A. H., Hardy, G. E. S. J., & Sivasithamparam, K. (2003). Fish emulsion as a food base for rhizobacteria promoting growth of radish (Raphanus sativus L. var. sativus) in a sandy soil. Plant and Soil, 252: 397-411.
Hatmanti, A. (2000). Pengenalan Bacillus spp. Balitbang lingkungan laut LIPI. Jakarta. 15(1):31-41.
Koolman, J., & Roehm, K. H. (2005). Color Atlas of Biochemistry. 2th ed. New. York: Georg Thieme Verlag. p. 164-162.
Lelliot, R. A., & Stead, D. E. (1987). Methods for the Diagnosis of Bacterial Disease of Plants. Blackwell Scientific Publication. London.
Miller, G., Beckwith, R., Fellbaum, C., Gross, D., & Miller, K. (1990).
WordNet: An on-line lexical
database. International Journal of Lexicography, 3: 235-244.
Pal, A., Chattopadhyay, A., & Paul, A. K. (2012). Diversity and antimicrobial spectrum of endophytic bacteria isolated from Paederia foetida L. International Journal of Current Pharmaceutical Research, 4 (3) : 123-127.
Seki, T., Chi, K. C., Hidetada, M., & Yasuji, O. (1978).
Deoxyribonucleaic Acid Homology and Taxonomy of the Genus Bacillus. International Journal of Systematic Bacteriology, 28(2):182-187.
Setiawati, M. R., Dedeh, H. A., Pujawati, S., & Ridha. (2009). Formulasi
Pupuk Hayati Bakteri Endofitik Penambat N2 dan Aplikasinya Untuk Meningkatkan Hasil
Tanaman Padi. Fakultas Pertanian UNPAD. Bandung. 7(1):1-7.
Tarigan, J. (1988), Pengantar
Mikrobiologi Umum. Departemen Pendidikan dan. Kebudayaan
Direktorat Jenderal Pendidikan
Tinggi. Jakarta.
ASIA OCEANIA BIOSCIENCE AND BIOTECHNOLOGY • 91
Discussion and feedback