Rapid Detection Of Methicillin Resistant Staphylococci Using Multiplex PCR With Boiling Method For DNA Isolation
on
Journal of Health Sciences and Medicine, Vol. 1 No. 2, September 2017
8
Rapid Detection Of Methicillin Resistant Staphylococci Using Multiplex PCR With Boiling Method For DNA Isolation
Ida Bagus Gede Adiguna Wibawa1, Agus Eka Darwinata1, Ni Nengah Dwi Fatmawati 1, Nyoman Sri Budayanti 1
-
1 Microbiology Departement, Faculty of Medicine, Udayana University, Bali, Indonesia
ABSTRACT.
Methicillin-Resistant Staphylococcus aureus (MRSA) is Staphylococcus aureus that has become insusceptible or resistant by methicillin antibiotic types. Rapid identification of MRSA is essential for early initiation of appropriate antimicrobial therapy. The aim of this research is to reduce the cost and time needed for multiplex PCR in rapid detection of MRSA by finding an alternative method for DNA which is boiling method. DNA isolation was performed with boiling method and kit commercial. The kit method takes time approximately 45 minutes while boiling takes only about 12 minutes. PCR result with boiling technique used in DNA isolation formed amplification bands of 16S rRNA, mecA, and nuc in MRSA and 16S rRNA and nuc in MSSA. Conclusion can be drawn that boiling method can be used as an alternative method for DNA extraction.
Keywords: MRSA, Multiplex PCR, DNA Isolation, Boiling Method
-
I. INTRODUCTION
Methicillin-Resistant Staphylococcus aureus (MRSA) is Staphylococcus aureus that has become insusceptible or resistant towards methicillin antibiotic types.1 MRSA has become resistant because of genetic changes that are caused by exposure of irrational antibiotic therapy. MRSA outbreak first occurred in Europe in the era of the 1960s. Then MRSA spread rapidly to various hospitals around the world. Therefore, the spread of
MRSA occurs between hospitals and raises the problem of infection in hospital, making it often called Healthcare-associated MRSA (HA-MRSA). Data shows that about 25% of isolates of S. aureus causes infections in hospitals in the United States is MRSA. The prevalence of MRSA in hospitals in the world ranged from 2-70% with the average rate of 20%. While the data or publications on MRSA in Indonesia is still very limited. In 2006, the prevalence stands at 23,5%.2,3,4 The rapid detection of methicillin-resistant staphylococci from bacteremic patients is essential for prompt and effective antimicrobial therapy. A rapid and simple multiplex PCR-based method is used to detect methicillin-resistant staphylococci with modification in DNA isolation method from kit to boiling method, for the purpose of cost and time reduction. Then, in this research Methicillin-Sensitive Staphyloccous aureus (MSSA) is used as a compared organism.
-
II. MATERIALS AND METHODS
-
A. Clinical Specimens
Staphylococcal isolate is an clinical isolate that had been identified in a clinical microbiology laboratorium as a part of a larger stock of spesimen. Staphylococcal isolate were indentified by standard methods, including gram stain, catalase test, oksidase test and cefoxitine disk.
-
B. DNA Isolation
DNA isolation was performed in two techniques which were boiling and kit commercial. For the kit commercial, highpure PCR template Preparation kit (Roche Diagnostic GmbH made in Germany) was used following the procedure to do the DNA isolation. In boiling method, the bacterial colony was placed in a tube. Then, 200μl Tris-EDTA buffer was used as a solution. The suspension was then boiled in 1000C water for 10 minutes. the suspension was centrifuged at 13,000 X g for 1 minute. The supernatant was later used for PCR testing. The kit method takes time approximately 45 minutes while boiling takes only about 12 minutes.
-
C. PCR Amplification
DNA was extracted from bacterial colony by using kit commercial and boiling method. Using template DNA, the polymerase chain reaction technique (PCR) was used to detect MecA, Nuc, and 16S genes. Primers used to amplify the screened genes are listed in Table 1. Multiplex PCR was performed using 20µl as total volume, consisting of 5μl of
mastermix (Kappa Biosystem based in wilmington), 0,2μl out of each of the 16sRNA and nuc primers, and 0,5μl of mecA primer. Thermocycling steps done in a biorad thermocycler were as follows: 2 minutes of predenaturation at 940C, followed by 30 seconds of denaturation at 940C, 30 cycles of annealing at 550C(each cycles takes 15 seconds) and 30 seconds of extension at 720. The PCR product were loaded on 2% agarose gel were analyzed by gel electrophoresis.
Table 1. Primers used for multiplex PCR of MRSA and MSSA5,6,7,8
|
Primer |
Sequence |
Product size (bp) |
Reference |
|
16S F |
AGAGTTTGATCATGGCTCAG |
768 | |
|
16S R |
GGACTACCAGGGTATCTAAT | ||
|
MecA F |
AAAATCGATGGTAAAGGTTGGC |
533 | |
|
MecA R |
AGTTCTGCAGTACCGGATTTGC | ||
|
Nuc F |
GCGATTGATGGTGATACGGTT |
270 | |
|
Nuc R |
AGCCAAGCCTTGACGAACTAAAGC |
-
III. RESULTS
The multiplex PCR was done for the simultaneous detection of the bacterial 16S rRNA, mecA, and nuc gene. Figure 1 is the result of multiplex PCR of MRSA and MSSA with different technique in DNA isolation. MRSA shows amplification of 16S rRNA, mecA and nuc gene while mssa shows only 16s rrna and nuc gene.
768 bp ----
533 bp ----
270 bp ----
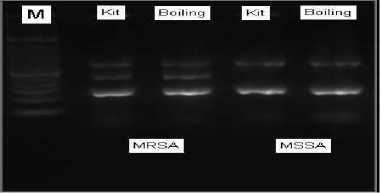
FIG. 1 Agarose gel showing PCR products of 16S at 798bp, mecA at 533 bp, and nuc at 270 bp. Lane 1 is marker. Lane 2 and 4 are the PCR results using kit commercial method for DNA isolation of MRSA and MSSA respectively. Lane 3 and 5 are the PCR results boiling method for DNA isolation of MRSA and MSSA respectively.
From lane 3 and lane 5, bands can be seen as result of multiplex PCR using boiling method. Therefore, it can be concluded that the boiling method can serve as an alternative method for DNA isolation and later then be processed in PCR.
-
IV. DISCUSSION
Before Multiplex PCR is done, there are several steps must be done, in the end, it takes a lot of time for all steps of PCR to be done. One of the steps that contribute to the long time needed for the PCR is the DNA isolation. Laboratories commonly use kit as the method to do DNA isolation, which approximately takes about 1 hour to conduct. By
finding an alternative method to do DNA isolation, the total time needed for the entire process of PCR can be significantly reduced. It is proved when DNA isolation was performed in two technique, which are Boiling method and kit commercial that takes time approximately 12 minutes and 45 minutes respectively. The result of PCR using boiling method for DNA isolation in the figure shows DNA amplification in the form of bands. As mentioned in the chapter before, conclusion can be drawn that boiling method can be used as an alternative method for DNA isolation. In addition, it can be seen from the time taken for each method of DNA isolation that 33 minutes is cut off from the time needed for kit method to the time needed for boiling method. In addition the advantage of saving time, the total cost needed for conducting DNA isolation is also significantly reduced by using boiling method compared to kit. Another advantage gained by using boiling method is the much fewer amount of DNA template needed for this method, as mentioned in the chapter before. Therefore, stocks of DNA template can be saved. The weakness of boiling method compared with kit is boiling method only get the crude DNA, while kit method can get pure DNA. Another study had been conducted by Louie et al for the rapid detection of MRSA using multiplex PCR with similar method in DNA isolation. In that study, 10-minute incubation in 370C water was done before the 10-minute boiling process, using Triton X-100 lysis buffer as the solution. That study presented that a rapid DNA isolation with multiplex PCR-based assay can be done within 3-hour time range.8
-
V. CONCLUSION
It is concluded that boiling method can be used as an alternative for DNA isolation in pure isolates, as done in this study, with fewer time and cost needed. Another study is suggested to be done to find whether boiling method can also be used as DNA isolation alternative in clinical specimens.
REFERENCES
-
[1] Whitby, M., M. L. McLaws, and G. Berry.2001. Risk of death from methi- cillin-resistant Staphylococcus aureus bacteraemia: a metaanalysis. Med. J. Aust. 175:264–267.
-
[2] Trakulsomboon S, Danchaivijitr S, Rongrungruang Y, Dhiraputra C,Susaemgrat W, Ito T, Hiramatsu K. First report of methicillin resistant Staphylococcus aureus reduced susceptibility to vancomycin in Thailand. J Clin Microbiol. 2001;39:591- 95.
-
[3] Jan M. Bell and John D. Turnidge. High Prevalence of OxacillinResistant Staphylococcus aureus Isolates from Hospitalized Patients in Asia-Pacific and South Africa: Results from SENTRY Antimicrobial Surveillance Program, 1998- 1999. Antimicrob Agents Chemother. 2002;46: 879-881.
-
[4] Vos MC, Ott A,Verbrugh HA. Successful Search-and-Destroy Policy [7]
for Methicillin-Resistant Staphylococcus aureus in The Netherlands J. Clin.
-
[5] Microbiol. 2005;43: 2034–2035
-
[6] acroix, J., K. Jarvi, S. Batra, D. Heritz, and M. Mittelman. 1996. [8]
PCR- based technique for the detection of bacteria in semen and urine. J. Micro- biol. Methods 26:61–71.
[9]
Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylo- cocci by polymerase chain reaction. J. Clin. Microbiol. 29:2240–2244.
Brakstad, O. G., K. Aasbakk, and J. Maeland. 1992. Detection of Staphylo- coccus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654–1660.
L. Louie. "Rapid Detection of Methicillin-Resistant Staphylococci from Blood Culture Bottles by Using a Multiplex PCR Assay", Journal of Clinical Microbiology, 08/01/2002
Discussion and feedback