MOLECULAR IDENTIFICATION OF FUNGI THE CAUSAL AGENT OF STRAWBERRY WILT DISEASE IN BALI
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 7 No. 2 • April 2020
eISSN: 2655-9994
pISSN: 2303-3371
https://doi.org/10.24843/IJBB.2020.v07.i02.p02
MOLECULAR IDENTIFICATION OF FUNGI THE CAUSAL AGENT OF STRAWBERRY WILT DISEASE IN BALI
Gusti Ngurah Alit Susanta Wirya*, I Wayan Diksa Gargita, and I Putu Sudiarta
Magister of Agriculture Biotechnology, Faculty of Agriculture, Udayana University Jl. PB. Sudirman, Denpasar, Bali 80232, Indonesia *Corresponding author: susantawirya@unud.ac.id
ABSTRACT
The development of strawberry farming in Bali experiencing some obstacles that cause a decline in production, such as wilting disease. The disease was reported caused by the fungi base on morphological recognition. There are two fungi were recognized caused the strawberry wilt disease in Bali, they are from genus Verticillium and Fusarium. More specific information about causal agent of wilt disease in strawberry especially in Bali is needed. The one accurate identification is done through the molecular approach by analyzing DNA that encode the ribosomal DNA (rDNA). The 18S rDNA, including the internal areas of transcribed spacers (ITS), ITS1 and ITS4 have been widely used in phylogenetic studies. The amplification results of this area produce bands in different sizes that can be used to identify fungal species. Based on that the identification of strawberry wilt disease using molecular analysis was conducted. The 542 bp of Internal Transcribed Spacer (ITS) DNA was successfully amplified using PCR with pairing primers ITS 1 (5-
TCCGTAGGTGAACCTGCGG-3’), and ITS 4 (5’-TCCTCCGCTTATTGATATGC-3’). The sequences of three isolates were successfully obtained through sequencing. Homology levels were tested between sequences and showed that Candi Kuning sequence and Gobleg sequence had 95% similarity with sequence of Fusarium oxysporum NRRL 13307 (U34571) from America. While Pancasari sequence have 94% similarity with sequence of Fusarium oxysporum NRRL 13307 (U34571) from America. Candi Kuning, Gobleg, and Pancasari sequences had the same 86% with sequence of Fusarium oxysporum isolate C34-294 Brazil (KJ439088) and had 89% similarity with sequence Fusarium oxysporum f.sp. fragariae China (KT833080). Homology levels were tested between sequences and showed that Candi Kuning sequence and Gobleg sequence had 95% similarity with sequence of Fusarium oxysporum NRRL 13307 (U34571) from America. While Pancasari sequence have 94% similarity with sequence of Fusarium oxysporum NRRL 13307 (U34571) from America. Candi Kuning, Gobleg, and Pancasari sequences had the same 86% with sequence of Fusarium oxysporum isolate C34-294 Brazil (KJ439088) and had 89% similarity with sequence Fusarium oxysporum f.sp. fragariae China (KT833080). Based on phylogeny analysis of Pancasari, Gobleg and Candi Kuning isolates were obtained in one group with Fusarium oxysporum identified in America and Brazil, and also in one group with Fusarium oxysporum f. sp. fragariae that identified in China.
Keywords: strawberry, wilt disease, pathogenicity, molecular
INTRODUCTION introduced in 1983 precisely in Candi
Strawberries entered Indonesia in the Kuning Village, Tabanan Regency, Bali and
1980s. Strawberries in Bali were first were developed to Bukit Catu Village,
Pemuteran Village and Batu Sesa Village Tabanan Regency, Bali. Strawberry is one of potential fruit because it contains many phytochemicals mainly phenolic compounds that are beneficial to human health (Hannum, 2004). Strawberries in Indonesia within a year can produce up to five times and peak production occurs from July to August depending on environmental conditions (Sukumalanandana and Verheij, 1997). However recently the production of strawberry fruit in Bali has been decreased, and one of the contributing factors is strawberry wilt diseases. The strawberry wilt diseases was reported caused by the fungi from the genus Verticillium (Sari et al., 2018). In contras some researcher reported that the strawberry wilt disease caused by Fusarium oxysporum in many countries such as Carolina (Williamson et al., 2012), Australia (Golzar et al., 2007), Serbia (Stankovic et al., 2014), and Spain (Arroyo et al., 2009). Based on the data there are two fungi were reported cased the strawberry wilt disease in Bali, they are from genus Verticillium and Fusarium. More specific information about causal agent of wilt disease in strawberry especially in Bali is needed. The one accurate identification method is done through the molecular approach by analyzing DNA that encode the ribosomal DNA (rDNA). Therefore on this study the identification of strawberry wilt
disease using molecular analysis was conducted.
MATERIALS AND METHODS Sample collection and fungi isolation
This research was started from April 2018 to June 2018. The wilting strawberry plants samples were collected at the center of strawberry plantations in Bali (Pancasari Village, Sukasada District, Buleleng Regency; Gobleg Village, Banjar District, Buleleng Regency and Candikuning Village, Baturiti District, Tabanan Regency) and Plant Disease Laboratory, Faculty of Agriculture, Udayana University. The isolations have done by cutting the infected parts (leaf, trunk or root) with the size about (1x1) cm2, then dipped it in alcohol for 2 minutes to remove contamination outside. Then, the pieces of plant were washed off by dipping it into the sterile water for 3 times, and then put it on Potato Dextrose Agar (PDA) medium which already contains chloramphenicol antibiotics (100mg/L) and grow on PDA medium which contained chloramphenicol antibiotics (100mg/L) (Ando et al., 2003).
Molecular identification of fungi on strawberry that infected wilt disease
Pure cultures were rejuvenated to the PDA medium. A disc (4 mm diameters) of mycelium was put at the middle of petri dish
that already contains PDA medium. After that, the cultures wereincubated for 24 hours and then used for DNA extraction.
DNA extraction and PCR
DNA extraction was done following a protocol based on ZR Fungal DNA Kit™ Catalog No. D6005 by ZYMO. The Genomic DNA extraction was conducted using ZR Fungal Bacteria DNA Kit (Zymo Research). The amplification odf DNA was conducted by PCR using MyTaq HS Red Mix that started by preparing the PCR master mix (9.5 µL ddH20, 12.5 µL MyTaq Red Mix [2x], 20 µmol primer ITS 1 [5-
TCCGTAGGTGAACCTGCGG-3’], 20
µmol primer ITS 4 [5’-
TCCTCCGCTTATTGATATGC-3’] (Singha et al., 2016), and I µL DNA template. The following cycling condition was used for PCR reaction: Initial denaturation at 94ºC for 60 s, then 35 cycles of 95ºC for 15 s, 52ºC for 15 s, 72ºC for 45 s, and 75ºC for 5 minutes. The PCR product was purified with ZymocleanÔ Gel DNA Recovery Kit (Zymo Research). Finally, bi-directional sequencing was conducted at Laboratory of Genetika Science in Kembangan Jakarta Barat.
Electrophoresis and sequencing
As much as 1 µL of PCR products were assessed by electrophoresis with 1%
TBE agarose gel. Electrophoresis was done for 30 minutes at 100 Volt. The electrophoresed DNA then was visualized with a UV transiluminator.
Sequence Analysis
DNA sequences were analyzed to make the alignment which then used to determine the level of homology or alignment with the sequence of ITS gene that has been published in GenBank within Program Basic Local Alignment Tool (BLAST) (NCBI 2014).
Phylogenetic Analysis
The analysis was continued with the phylogenetic analysis by using software: ChromasPro, Molecular Evolutionary Genetics Analysis (MEGA 6.06), PAUP 4.0, and TreeGraph2.0.
RESULT AND DISCUSSION
Molecular Identification of Fungi on Strawberry that Infected Wilt Disease
Fungal cultures from Pancasari Village, Gobleg Village, and Candikuning Village, were successfully obtained, reproduced and identified morphologically based on macroscopic character (Fig. 1). Therefore the culture of the fungi can be used in the molecular identification phase.
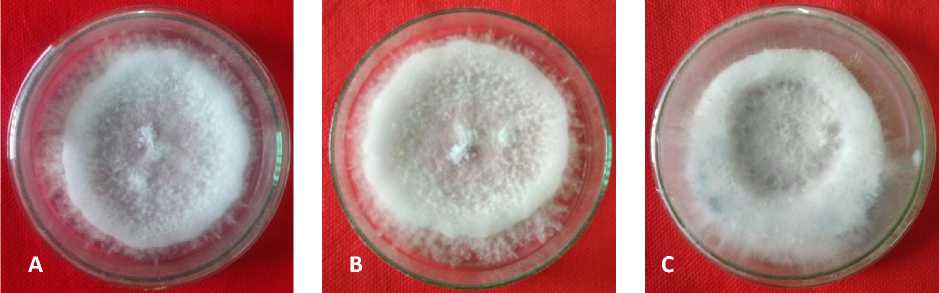
Fig. 1. Pure culture 7 days-old on PDA medium of A) Pancasari isolate, B) Gobleg isolate, and C) Candi Kuning isolate that isolated from strawberry plant on pathogenicity test
According to Sari et al., (2018), the decrease in strawberry production due to infection by fungi was very high. The decrease in strawberry production that occurred in Pancasari Village reached 95% and in Candi Kuning Village it reached 85%. The decrease in plant production due to the attack of Fusarium fungus is very dependent on the attack intensity, so the decline in production in each region will be different. This is because, Fusarium fungus is strongly influenced by environmental conditions. When the environmental conditions are suitable, the fungus will be able to reduce very high plant production. Environmental conditions that are not suitable for the development of Fusarium fungi, cause a decrease in plant production also decreases (Nagy et al., 2006). It was supported by Melysa et al. (2013), that plant infections carried out by Fusarium sp. affected by humid weather conditions and plant age.
Younger plant ages tend to be more susceptible to disease than adults plant. Disease caused by Fusarium sp. easier to attack plants in the high rainy season.
In addition to environmental factors and plant age, the factor of varieties resistance may also affect the decrease in strawberry plant production. Considering, the development of strawberries in Bali is still depend on strawberry seedlings that taken from the former brood-stock. Broodstock varieties of developed in strawberry production centers in Bali, especially in Pancasari Village, Gobleg Village and Candi Kuning Village are mixed varieties. Each variety for strawberry seedlings that taken from the previous brood-stock is certainly unknown. Therefore, resistant varieties and non-resistant varieties were mixed at a strawberry plantation that were mainly developed. The number of resistant and nonresistant varieties in each region certainly
will not be the same, so the decreasing in strawberry production in each region will possibly different. It was compatible with the statement by Melysa et al. (2013), that strawberry plants of California varieties were more sensitive to disease. The results of the analysis of the disease intensity of the in vivo test on California varieties and Santung varieties can be seen that California varieties were more sensitive than Santung varieties. The intensity of pathogen attacks on California varieties were higher than Santung varieties.
Another research such as in Australia (Golzar et al., 2007; Fang et al., 2012), United States (Koike et al., 2009, 2013), Spain (Arroyo et al., 2009), Korea (Nagarajan et al., 2004) and China (Zhao et al., 2009) stated that F. oxysporum f. sp. fragariae has recently been considered as one of the most serious fungal threats to strawberry cultivation and it can also
threaten strawberry fruit yield and transplant production in Bali.
Identification of pathogenic fungi that cause strawberry wilt disease was carried out molecularly by amplifying the Internal Transcribed Spacer (ITS) area by using two primers, namely ITS 1 and ITS 4 rDNA (ribosomal DNA). One of the molecular analysis that have been developed is using ribosomal DNA sequences in the Internal Transcribed Spacer area (Druzhirina and Kubicek, 2005). DNA amplification in this molecular analysis was carried out to multiply gene fragments located in the Internal Transcribed Spacer area. The DNS fragment size of the amplification target with the primer used was in accordance with the findings stated by White et al. (1990)The PCR product obtained and electrophoresed, showed the DNA band that appeared. The amplified DNA band showed the base length are 542 bp (Gobleg), 542 bp (Candi Kuning), and 544 bp (Pancasari) (Fig. 2).
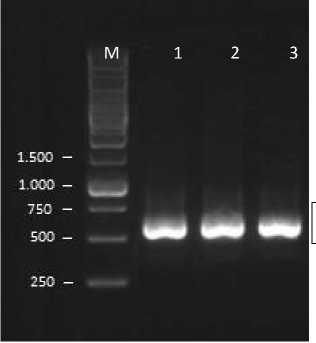
542
Fig. 2. 1 µL PCR Products were assessed by electrophoresis with 1% TBE agarose showed bands DNA 1st). Lane 1 is 542 bp band (Gobleg), 2nd). Lane 2 is 542 bp band (Candi Kuning), 3rd). Lane 3 is 544 bp band (Pancasari), and M= DNA marker 1kb
The appearance of DNA bands is an important information that shows that PCR has been successfully carried out on the DNA sample that was believed as Fusarium oxysporum. However, these results have not been able to ascertain the truth about whether the amplified is truly DNA from Fusarium oxysporum because the primer used is a
universal primer. Although, in accordance with the statement from Singha et al. (2016), the Fusarium oxysporum species that amplified using ITS1 and ITS4 primers produced 380-620 bp sized bands. Therefore, sequencing was carried out to ensure the correctness of the information needed in this study.
Table 1. Homology of the fungus that causes strawberry wilt disease in Bali with its homologous sequence in GeneBank
|
No. |
Isolate 1 2 3 4 5 6 7 8 9 10 11 12 13 |
|
1 |
Candi Kuning |
|
2 |
Gobleg 100% ID |
|
3 |
Pancasari 100% 100% ID |
|
4 |
U34571 95% 95% 94% ID |
|
5 |
KJ439088 86% 86% 86% 91% ID |
|
6 |
KT833080 89% 89% 89% 94% 96% ID |
|
7 |
LN809075 76% 76% 76% 79% 78% 80% ID |
|
8 |
KR071709 36% 36% 36% 37% 40% 39% 37% ID |
|
9 |
KR071713 36% 36% 36% 37% 40% 39% 38% 94% ID |
|
10 |
KC874687 37% 37% 37% 38% 37% 37% 36% 85% 84% ID |
|
11 |
KF641021 36% 36% 36% 37% 35% 35% 34% 80% 81% 86% ID |
|
12 |
KM487196 34% 34% 34% 36% 39% 38% 36% 90% 91% 82% 77% ID |
|
13 |
DQ266147 28% 28% 28% 29% 32% 31% 30% 38% 38% 35% 32% 37% ID |
Note:
Candi Kuning: Candi Kuning Isolate; Gobleg: Gobleg Isolate; Pancasari: Pancasari Isolate; U34571: U34571 1 Fusarium oxysporum NRRL 13307 America; KJ439088: KJ439088 1 Fusarium oxysporum isolate C34-294 Brazil; KT833080: KT833080 1 Fusarium oxysporum f.sp. fragariae China; LN809075: LN809075 1 Fusarium solani Isolate 1111TES31E1; KR071709: KR071709 1 Fusarium verticillioides Strain CBS 108922; KR071713: KR071713 1 Fusarium napiforme Strain CBS 674-94; FFK: KC874687 1 Fusarium fujikuroi Isolate F148N4; KF641021: KF641021 1 Fusarium nygamai Isolate M42; KM487196: KM487196 1 Fusarium subglutinans strain MUCL 43485; DQ266147: DQ266147 1 Verticillium alboatrum Isolate V48I.
The similarity levels of samples sequences with sequences in GeneBank were showed in Table 1. Homology levels were tested between sequences and showed that
Candi Kuning sequence and Gobleg sequence had 95% similarity with sequence of Fusarium oxysporum NRRL 13307 (U34571) from America. While Pancasari
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 7 No. 2 • April 2020
sequence have 94% similarity with sequence of Fusarium oxysporum NRRL 13307 (U34571) from America. Candi Kuning, Gobleg, and Pancasari sequences had the same 86% with sequence of Fusarium
eISSN: 2655-9994 pISSN: 2303-3371 https://doi.org/10.24843/IJBB.2020.v07.i02.p02
oxysporum isolate C34-294 Brazil (KJ439088) and had 89% similarity with sequence Fusarium oxysporum f.sp. fragariae China (KT833080).
Table 2. The genetic distance of the fungus that causes strawberry wilt disease in Bali with its homologous sequence in GeneBank
|
No. |
Isolate |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
|
1 |
Candi Kuning |
ID | ||||||||||||
|
2 |
Gobleg |
0.000 |
ID | |||||||||||
|
3 |
Pancasari |
0.000 |
0.000 |
ID | ||||||||||
|
4 |
U34571 |
0.005 |
0.005 |
0.005 |
ID | |||||||||
|
5 |
KJ439088 |
0.002 |
0.002 |
0.002 |
0.002 |
ID | ||||||||
|
6 |
KT833080 |
0.002 |
0.002 |
0.002 |
0.002 |
0.000 |
ID | |||||||
|
7 |
LN809075 |
0.142 |
0.142 |
0.142 |
0.142 |
0.140 |
0.140 |
ID | ||||||
|
8 |
KR071709 |
0.578 |
0.578 |
0.578 |
0.573 |
0.576 |
0.576 |
0.607 |
ID | |||||
|
9 |
KR071713 |
0.578 |
0.578 |
0.578 |
0.573 |
0.576 |
0.576 |
0.602 |
0.055 |
ID | ||||
|
10 |
KC874687 |
0.573 |
0.573 |
0.573 |
0.573 |
0.571 |
0.571 |
0.604 |
0.057 |
0.071 |
ID | |||
|
11 |
KF641021 |
0.581 |
0.581 |
0.581 |
0.581 |
0.578 |
0.578 |
0.618 |
0.062 |
0.050 |
0.066 |
ID | ||
|
12 |
KM487196 |
0.576 |
0.576 |
0.576 |
0.571 |
0.573 |
0.573 |
0.607 |
0.059 |
0.052 |
0.076 |
0.066 |
ID | |
|
13 |
DQ266147 |
0.637 |
0.637 |
0.637 |
0.642 |
0.640 |
0.640 |
0.645 |
0.590 |
0.583 |
0.581 |
0.592 |
0.597 |
ID |
Note: Candi Kuning: Candi Kuning Isolate; Gobleg: Gobleg Isolate; Pancasari: Pancasari
Isolate; U34571: U34571 1 Fusarium oxysporum NRRL 13307 America; KJ439088: KJ439088 1 Fusarium oxysporum isolate C34-294 Brazil; KT833080: KT833080 1 Fusarium oxysporum f.sp. fragariae China; LN809075: LN809075 1 Fusarium solani Isolate 1111TES31E1; KR071709: KR071709 1 Fusarium verticillioides Strain CBS
108922; KR071713: KR071713 1 Fusarium napiforme Strain CBS 674-94; FFK:
KC874687 1 Fusarium fujikuroi Isolate F148N4; KF641021: KF641021 1 Fusarium
nygamai Isolate M42; KM487196: KM487196 1 Fusarium subglutinans strain MUCL 43485; DQ266147: DQ266147 1 Verticillium alboatrum Isolate V48I.
Based on the data in Table 2, it showed that Candi Kuning sequence, Gobleg sequence and Pancasari sequence have the lowest genetic distance value with sequence of Fusarium oxysporum isolate C34-294 Brazil and sequence of Fusarium oxysporum f.sp. fragariae China are 0,002.
The genetic distance value of Candi Kuning sequence, Gobleg sequence and Pancasari sequence with sequence of Fusarium oxysporum NRRL 13307 America are 0,005. Genetic distance with a value of 0,002-0,005 are low. Low genetic distance was observed between the Candi Kuning
sequence, the Gobleg sequence and the Pancasari sequence with the sequence of Fusarium oxysporum isolate C34-294 Brazil, the Fusarium oxysporum f.sp. fragariae China, and sequence of Fusarium oxysporum NRRL 13307 America. This shows that the similarity level of the fifth sequences were very high. The genetic distance value of Candi Kuning sequence, Gobleg sequence
and Pancasari sequence with sequence of Verticillium alboatrum isolate V48I as outgroup is 0,637. High genetic distance values indicate that the similarity between the two genes is very low. Verticillium alboatrum is used as an outgroup because it is a pathogenic fungus in strawberry plants and has characteristics similar to Fusarium when identified morphologically.
100
100
52.4
100
69.6
91.2
50.8
-CandiKuning ◄—
— Gobleg -→-
-Pancasari →-
— U34571 1 Fusarium oxysporum NRRL 13307 America — KJ439088 1 Fusarium oxysporum isolate C34 294 Brazil — KT833080 1 Fusarium oxysporum f sp fragariae China — LN809075 1 Fusarium solani isolate 1111TES31E1 — KR071709 1 Fusarium Verticillioidesstrain CBS 108922 j—KR071713 1 Fusarium napiforme strain CBS 674 94 I—KF641021 1 Fusarium nygamai isolate M42
— KM487196 1 Fusariumsubglutinansstrain MUCL 43485 — KC874687 1 Fusarium fujikuroi isolate F148N4
----DQ266147 1 Verticillium alboatrum isolate V48l
I.........In.......I
0.0 100.0
Fig. 3. Phylogenetic tree arranged based on DNA composition of Candi Kuning, Gobleg and Pancasari isolates with the Maximum Parsimony method. The number in the branch is the percentage of the level of trust in the group
Phylogeny tree which show the proximity of the Pancasari, Gobleg and Candi Kuning fungi isolates to several other species based on bootstrap 1,000 times replication are presented in Fig. 3. Based on the phylogeny analysis of Pancasari, Gobleg
and Candi Kuning isolates were obtained in one group with Fusarium oxysporum that identified in America and Brazil, and also in one group with Fusarium oxysporum f.sp. fragariae that identified in China. This is consistent with the statement of Dharmayanti
(2011) that in a phylogenetic topology branches becomes significant if the value of the data set is > 70%.
CONCLUSION
Based on molecular analysis of DNA sequences from Pancasari isolate, Gobleg isolate and Candi Kuning isolate are fungi from the genus of Fusarium and the species are Fusarium oxysporum that identified in America and Brazil, and also the species are Fusarium oxysporum f.sp. fragariae that identified in China.
REFERENCES
Ando, K., C. Nakhashima, J-Y. Park, and M.
Otoguro. 2003. Workshop on Isolation Methods of Microbes.
Biotechnology Center-NITE &
Research and Development Center for Biotechnology-LIPI, Cibinong: 24--26 Juni 2003.
Arroyo, F.T., Y. Llergo, A. Aguado and F.
Romero. 2009. First Report of Fusarium Wilt of Strawberry Caused by Fusarium oxysporum in Spain. Plant Disease. 93: 323.
Druzhirina and Kubicek C.P. 2005. Species concepts and biodiversity in Trichoderma and Hypocrea: from aggregate species to species clusters? J. Zhejiang. Univ. Sci. B 6: 100-112.
Fang, X., J. Kuo, M. You, P.M.
Finnegan, and M.J. Barbetti.
2012. Comparative root colonisation of strawberry cultivars Camarosa and Festival by Fusarium oxysporum f. sp. fragariae. Plant Soil 358:75-89.
Golzar, H., D. Phillips and S. Mack. 2007.
Occurrence of strawberry root and crown rot in Western Australia.
Australasian Plant Disease Notes. 2: 145-147.
Hannum, S. M. 2004. Potential impact of strawberries on human health. Crit. Rev. Food Sci. Nutr. 44:1-17.
Koike, S.T.,S.C. Kirkpatrick and T.R. Gordon. 2009. First Report of Fusarium Wilt of Strawberry Caused by Fusarium oxysporum in California. Plant Disease. 93: 1007.
Melysa, D.E., M. N. Fajri, and Yunimar. 2013. Potensi Trichoderma sp. Sebagai Agen Pengendali Fusarium sp. Patogen Tanaman Strawberry (Fragaria sp.). J. Biotrop. 1 (4):177-181.
Nagarajan, G., S. W. Kang, M. H. Nam, J. Y. Song, S. J. Yoo, and H. G. Kim. 2006. Characterization of Fusarium oxysporum f. sp. fragariae based on vegetative compatibility group, random amplified polymorphic DNA and pathogenicity. P. Pathol. 22:222229.
Nagy, E., H. Voichita, and R. Kadar. 2006. The influence of Fusarium ear infection on the maize yield and quality (Transylvania-Romania).
Commun. Agric. Appl. Biol. Sci. 71:1147-1150.
Sari, D. V., Wirya, G.N.A.S., Sudiarta, I P. 2018. Identification of the causes of lute diseases in strawberry plants (Fragaria sp.) in Pancasari Village and Control Potential by Microbial Antagonists. J. of Trop. Agroecotech 7(1):103-112. Indonesia.
Singha, I.M., Yelena K,, B.G. Unni, J. Das, and M.C. Kalita. 2016. Identification and characterization of Fusarium sp. using ITS and RAPD causing fusarium wilt of tomato isolated from Assam, North East India. J. of Gen. Eng. and Biotechology,. . 14(1): 99105.
Stankovic, I., D. Ristic, A. Vucurovic, K. Milojevic, D. Nikolic, B. Krstic and A. Bulajic. 2014. First Report of Fusarium Wilt of Strawberry Caused
by Fusarium oxysporum in Serbia. Plant Disease 98(10):140722152922009.
Sukumalanandana, C. and E. W. M. Verheij. 1997. Fragaria x
ananassa (Duchesne). In E.W.M. Verheij and R.E. Coronel (Eds.). Edible Fruits and Nuts. Prosea Plant Resources of South-East Asia. Bogor, Indonesia.
Williamson, M., F. Ortuño D., Schnabel G. 2012. First Report of Fusarium Wilt of Strawberry Caused by Fusarium oxysporum in South Carolina. Plant Disease 96: 911.
White, T.J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M., A, Gelfand, D., H, Sninsky, J., J, White, T., J (Eds.), PCR protocols, a guide to methods and applications. Academic Press, Inc, California, pp. 315-322.
Zhao, X. S., W. C. Zhen, Y. Z. Qi, X. J. Liu, B. Z. Yin. 2009. Coordinated effects of root autotoxic substances and Fusarium oxysporum Schl. f. sp. fragariae on the growth and replant disease of strawberry. Front. of Agric. in China 3:34-39.
FACULTY OF AGRICULTURE, UDAYANA UNIVERSITY • 73
Discussion and feedback