IDENTIFICATION OF CITRUS WHITEFLY, HOST OF ENTOMOPATHOGENIC FUNGI (ASCHERSONIA PLACENTA) IN BALI INDONESIA
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 7 No. 2 • April 2020
eISSN: 2655-9994
pISSN: 2303-3371
https://doi.org/10.24843/IJBB.2020.v07.i02.p01
IDENTIFICATION OF CITRUS WHITEFLY, HOST OF ENTOMOPATHOGENIC FUNGI (ASCHERSONIA PLACENTA) IN BALI INDONESIA
Ni Putu Merthaningsih, I Putu Sudiarta*, and Gusti Ngurah Alit Susanta Wirya
Magister of Agriculture Biotechnology, Faculty of Agriculture, Udayana University Jl. PB. Sudirman, Denpasar, Bali 80232, Indonesia
*Corresponding author: putusudiarta@unud.ac.id
ABSTRACT
One of the pests of citrus is whitefly that causes damage directly or/and indirectly to the citrus production. To control whitefly the farmer usually use chemical insecticide, however the utilization of chemical insecticide has been reported to haves many negative effect. To minimize the utilization of chemical insecticide, the environmentally friendly method is needed. One of the method is to utilize the natural enemies. Natural enemies are including, parasitiod, predator as well as insect pathogen (entomopathogen). In 2017 entomopathogenic fungi Aschersonia placenta was found to be associated with citrus whitefly in Bali Indonesia. However the species of whitefly has not been identified. In this research the identification of whitefly, the host insect of A. placenta was conducted based on morphological and molecular identification. Morphological identification of whitefly use puparial stage, started with sample preparation by Slide Mounting Protocol. The target of mitochondrial cytochrome c oxidase subunit I (mtCOI) gen was successfully amplified (700 bp) by PCR using forward primer LCO 5'GGTCAACAAATCATAAAGATATTGG3' and reverse primer HCO 5'TAAACTTCAGGGTGACCAAAAAATCA3'. The phylogenetic analysis using software ChromasPRO, Molecular Evolutionary Genetics Analysis (MEGA 5.05), PAUP, BioEdit, and TreeGraph2 was conducted. The result shows that the mtCOI sequence of P. minei from Bali (LC491421) has the highest percentage among others with MK421974 P. minei (score homology 96%). The morphological recognition and sequence analysis show that the species of citrus whitefly is Paraleyrodes minei.
Keywords: whitefly, host insect, citrus, Aschersonia placenta, Paraleyrodes minei
INTRODUCTION
Whitefly is a polyphagous insect that has a very broad host distribution, these insects are widespread in the tropics and subtropics. Whiteflies can cause damage directly or indirectly. Direct damage as a food activity, such as (1) Closure of stomata by honey dew released by nymphs and soot dew which grows on the honey dew layer; (2) Chlorotic spots on leaves resulting in
damage to tissue due to stylet puncture, (3) formation of anthocyanin pigments; and (4) Leaves fall so that can inhibit plant growth (De Barro 1995; Setiawati et al., 2007). Indirect damage is generally whiteflies is a vector of viruses that can cause disease in plants. The abundance of whiteflies infestation has been reported in citrus plant (Syafitri et al., 2017), reported causing a decrease in yield due to increased whitefly
infestation. The control of whiteflies is currently using insecticides that are harmful to the environment and human health. One approach that is more environmentally friendly is to use natural enemies. One of the natural enemies of the whiteflies is the entomopathogenic fungus. The fungus is Aschersonia placenta, the species was found associated with whiteflies in citrus plant in Bali Indonesia (Sudiarta, et al., 2017; Suputra, et al., 2019). The species of whitefly as the host of A. placenta has not been identified, even though the
information is very important for basic science and applied science. The whitefly on citrus plant in Bali was predicted as Dialeurodes citri from morphological recognise from field, however the small body and the similarity morphological from one and other whitefly make difficult to accurate identification. Therefore on this study, the identification with morphological and molecular characteristic of whitefly was conducted.
MATERIALS AND METHODS Morphological Identification of Whiteflies
Samples were taken from the results of field surveys, where the symptoms of Aschersonia sp. found, in various districts in Bali (Gianyar, Bangli, Tabanan and Buleleng). Sampling was conducted by
taking nymphs of whitefly, which is usually found under the leaf. Morphological identification of whiteflies using puparial stage, begins with sample preparation by Slide Mounting Protocol (modified from Wilkey 1962 in Gregory et al., 2005). Specimens are identified using a microscope by comparing insect morphology by matching each part and compared with the literature than by photographing preparations using
microscopic cameras which are then identified by matching the literature. Morphological identification of whitefly using literature “An Identification Guide To The Whiteflies (Hemiptera: Aleyrodidae) of The Southeastern United States” by Gregory et al., 2005.
Molecular Identification of Whiteflies DNA Extraction and amplification
Protocol based on ZR Insect DNA Kit™ Catalog No. D6016 by ZYMO RESEARCH. PCR amplification using the primary pair of M-COI, LCO Primer: 5' GGTCAACAAATCATAAAGATATTGG 3' as forward primer and HCO Primer: 5' TAAACTTCAGGGTGACCAAAAAATC A 3' as reverse primer.
Electrophoresis
The 1 μl PCR product (plus 2 μl of loading dye) was electrophoresed in agarose
gel. Electrophoresis was carried out for 30 minutes at 100 volts. The DNA that has been electrophoresed is then visualized with a translator UV.
Sequencing
Sequencing was held at Laboratory of Genetika Science Indonesia for sequence of nucleotides. The sequence results then were analyzed to make the alignment then was used to determine the level of homology or alignment with sequence gen mitochondrial cytochrome c oxidase subunit I (mtCOI) gen whiteflies that have been published in GenBank with the Basic Local Alignment Tool (BLAST) Program (NCBI 2014).
Phylogenetic Analysis
The sequence of nucleotide data then followed by phylogenetic analysis using software ChromasPRO, Molecular Evolutionary Genetics Analysis (MEGA 5.05), PAUP, BioEdit, and TreeGraph2.
RESULTS AND DISCUSSION Morphological analysis
Morphological identification was conducted using nymph stage sample. Full body shape of whitefly was found in citrus plant is follows (Fig. 1). Specific morphological features that are very different from other whitefly species and can be used to identify whitefly as body shape, thoracic legs with claws, compound pore, dorsal disc, and vasiform (Fig. 2).

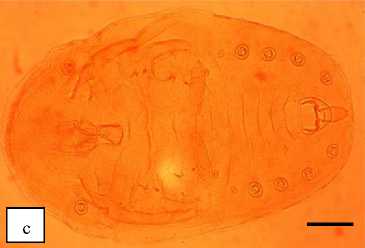
Fig. 1. The Full body of Paraleyrodes minei. a) Imago of P. minei; b) Nymph of P. minei; c) Full body shape of P. minei after preparation (magnification 10x using stereo microscope. Bar: 100 µm)p
This species has morphological characteristics, among others: in exuvia there are five pairs of abdominal compound pores that are shaped like a crown or splinal.
Compound pores with a series of rod-like processes in a ring. One pair on the head and four pairs on the subdorsal part of segments five to eight.
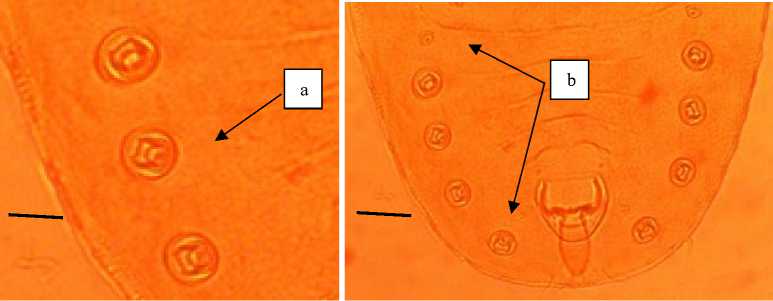
Fig. 2. Paraleyrodes minei species after preparation. a) Compound pores with rod-like processes. b) five pairs of abdominal compound pores that are shaped like a crown or splinal. Magnification 40x using stereo microscope. Bar: 100 µm
In addition there are two pairs of reduced pores in the anterior abdominal section and at the tip of the lingula there are two pairs of seta. Nurjanah (2016) stated that whitefly having abdominal compound pores (2 pairs of reduced size in the anterior portion), there is a discal pore between the abdominal compound pores VIII with vasiform orifice, known as Paraleyrodes minei. Basen on the data the morphological character of citrus whitefly from Bali Indonesia is very similar with morphology of P. minei.
Molecular analysis
Sequence Homology, Genetic Distance and Phylogenetic Analyze of Paraleyrodes minei
Molecular analysis was conducted to support the morphological identification results. Molecular analysis of citrus whitefly sample has been carried out successfully in several stages. The results of whitefly DNA extraction was then amplified using PCR with forward primer LCO and reverse primer HCO for the amplification of mitochondrial cytochrome c oxidase subunit I (mtCOI) (Folmer et al., 1994). The product from PCR was then electrophoresed and sequenced then the data obtained was analysed to get the desired information. The PCR product obtained and electrophoresed
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 7 No. 2 • April 2020
showed the DNA band that appeared. The amplified DNA band showed a base length 700 bp. The emergence of DNA bands shows that PCR has been successfully carried out on the whitefly DNA. This PCR had use universal primers, therefore sequencing was needed to ensure the correctness of the information. The similarity of sequence between P. minei
eISSN: 2655-9994 pISSN: 2303-3371 https://doi.org/10.24843/IJBB.2020.v07.i02.p01
species with the order from GenBank can be seen in Table 1.
The sequence alignment results using clustalW based on table 1, it can be seen that the P. minei sequence has the highest percentage among others with MK421974 P. minei (score homology 96%). With other species, which are still in one family, the homology is quite low, ranging from 37-40%.
Table 1. Similarity of Paraleyrodes minei with homologous sequence in GenBank based on COI
|
No. |
Accssion Number |
Homologv | |||||||
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 | ||
|
1 |
LC491421 |
ID | |||||||
|
2 |
MK421974 |
96% |
ID | ||||||
|
3 |
KP032214 |
42% |
42% |
ID | |||||
|
4 |
MK490947 |
42% |
42% |
100% |
ID | ||||
|
5 |
KX925199 |
42% |
42% |
96% |
96% |
ID | |||
|
6 |
MK343480 |
42% |
42% |
80% |
80% |
78% |
ID | ||
|
7 |
KY841808 |
39% |
38% |
75% |
75% |
73% |
73% |
ID | |
|
8 |
KP418776 |
22% |
21% |
26% |
26% |
26% |
26% |
27% |
ID |
Note: LC491421 Paraleyrodes minei (sample); MK421974 Paraleyrodes_minei; KP032214 Aleurodicinae sp.; MK490947 Paraleyrodes minei; KX925199 Tetraleurodes
perseae; MK343480 Paraleyrodes bondari; KY841808 Aleyrodidae sp.; AB470712 Acyrthosiphon pisum; KP418776 Acaudaleyrodes rachipora.
The results of the phylogeny analysis on the P. minei (Fig. 3) divide the sequence into several groups. P. minei sequences form an own separate group. P.
minei sample then formed a wider branching with group MK421974 P. minei which indicates that both sample are same species.
go KP032214_Aleurodicinae
93 MK490947_Paraleyrodes_minei
. ∣∙ KX925199_Tetraleurodes_perseae
----MK343480 Paraleyrodesboπdari
-------KY841808_Aleyrodidae_sp.
LC491421_Paraleyrodes_minei
100 ΓMK421974 Paraleyrodes-minei
------------------------------------∙ KP418776.1
Fig. 3. Phylogenetic tree arranged based on DNA composition of Paraleyrodes minei with the Maximum Parsimony method. The number in the branch is the percentage of the level of trust in the group
Dialeurodes citri is the most common pest that attacking citrus plants. There is a suspicion that A. placenta which infects whitefly on citrus plants in Bali is Dialeurodes citri. After morphological and molecular identification, the result show that the host insect of A. placenta in several region in Bali is a Paraleyrodes minei.
CONCLUSION
The citrus whitefly host insect of Aschersonia placenta from Bali Indonesia was identified as Paraleyrodes minei, based on the morphological and molecular characteristics.
ACKNOWLAGEMENTS
The authors wish to express their gratitude to the Research and Community Services Institution, Udayana University,
which funded the research on scheme of “Penelitian Unggulan Program Studi 2020”.
REFERENCES
De Barro, P.J. (1995). Bemisia tabaci Biotype B, a Review of its Biology, Distribution and Contro. CSIRO Division Entomology Technical Paper. 36:1-58.
http://hdl.handle.net/102.100.100/235 540?index=1
Folmer O., Black M., Hoeh W., Lutz R., and Vrijenhoek R.. (1994). DNA primers for amplification of mitochondrial Cytochrome C oxidase subunit I from diverse metazoan invertebrates.
Molecular Marine Biology and
Biotechnology 3(5), 294-299.
https://www.ncbi.nlm.nih.gov/pubme d/7881515
Gregory S. Hodges and Gregory A. Evans. (2005). An Identification Guide to The Whiteflies (Hemiptera:
Aleyrodidae) of The Southeastern United States . Florida Entomologist 88(4): 518-534.
http://dx.doi.org/10.1653/0015-
4040(2005)88[518:AIGTTW]2.0.CO;
2
Nurjanah. (2012). Keanekaragaman Spesies Kutukebul (Hemiptera: Aleyrodidae) di Taman Buah Mekarsari Kabupaten Bogor [skripsi]. Bogor (ID): IPB.
Setiawati, W., Udiarto B.K., dan Gunaeni N.
-
(2007) . Preferensi Beberapa Varietas
Tomat dan Pola Infestasi Hama Kutu Kebul serta Pengaruhnya terhadap Intensitas Serangan Virus Kuning. J. Hort, 17(4):374-386.
DOI: http://dx.doi.org/10.21082/jhort. v17n4.2007.p%25p
Sudiarta I P., Suputra, I P.W., Wirya, G.N.A.S. (2019). New Report of Insect Pathogenic Fungi (Aschersonia sp.) of Citrus Whitefly (Dialeurodes sp.) In Bali Indonesia. Agricultural and Veterinary Science, 3(1). http://jomardpublishing.com/UploadF iles/Files/journals/RV/V3N1/Sudiarta %20et%20al.pdf
Suputra I P. W., Sudiarta I P., Wirya, G. N.
-
A. S., and Sumiartha I K. (2019).
New Report of Distribution of Entomopathogenic Fungi Aschersonia sp. Infected Whitefly in Bali. AGROTROP, 9 (2): 188 – 196.
https://doi.org/10.24843/AJoAS.2019. v09.i02.p10
Syafitri D.D., Hafiz J.J., Salbiah, F.D. (2017). Kelimpahan Hama Kutu pada Tanaman Jeruk Siam (Citrus nobilis Lour.) di Desa Kuok Kecamatan Kuok Kabupaten Kampar Provinsi Riau. JOM Faperta, (4):1. https://jom.unri.ac.id/index.php/JOM
FAPERTA/article/view/1631
FACULTY OF AGRICULTURE, UDAYANA UNIVERSITY • 63
Discussion and feedback