PCR FINGERPRINTING OF DIVERSE GENOMES FROM BACTERIAL STRAINS USING UNIVERSAL RICE PRIMER (URP)
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 6 No. 1 • September 2018
ISSN: 2303-3371
https://doi.org/10.24843/IJBB.2018.v06.i01.p06
PCR FINGERPRINTING OF DIVERSE GENOMES FROM BACTERIAL STRAINS USING UNIVERSAL RICE PRIMER (URP)
Hee Wan Kang1, 2, 3*
1Department of Horticultural life science, Hankyong National University, Ansung 17579, Korea 2Institute of Genetic Engineering, Hankyong National University, Anseong 17579, Korea 3JK BioTech.Co Ltd. Hankyong National University, Ansung 17579, Korea
*Corresponding author: kanghw2@hknu.ac.com
ABSTRACT
Twenty primers of 20 mer referred to universal rice primer (URP) were developed from a repetitive sequence of rice genome. URP-PCR protocol employed stringent PCR with high annealing temperature throughout the thermo-cycling reaction, giving high reproducibility. Under the PCR condition, each single URP primer produced characteristic fingerprints from diverse genomes of bacterial species. The universal application of URP-PCR was demonstrated by applying it to 24 strains from Pectobacterium carotovoum subsp. carotovorum, 41 Agrobacterium vitis strains, 3 Xanthomonas spp. 5 Pseudomonas spp, Rhizobium sp. plant pathogenic bacteria, human and animal pathogenic bacterial strains including 6 Escherichia coli, 4 Salmonella spp., 7 Mycobacterium spp and 3 Blucella abortus strains. In addition, thermophilic bacteria were randomly isolated form high temperature compost and their URP-PCR polymorphisms were characterized with genetic relatedness. PCR approach using URP primers will be useful for studying DNA diversity of diverse prokaryotic genomes, especially at inter- and intra species levels.
Keywords: Diverse bacterial strains, PCR polymorphism, Universal Rice Primers
INTRODUCTION
The classic taxonomical methods of bacteria, relies on fatty acids, nutritional composition, and biochemical properties and so on (Garrity et al., 2002). However, it has been pointed out that microorganisms tend to lead to other consequences due to environmental effects, and that their shorter
generation and higher rates of metamorphosis have been a limiting factor in the bacterial classification system. The sequence of 16S rDNA was adopted as a new classification tool of the bacterial class (Chang et al., 1997), providing an opportunity for the revision of bacterial taxonomy to take place. In particular, inter transcribed spacer (ITS)-16S
rDNA sequence region is widely used as a species-specific identification of bacterial species (Chang et al., 1997, Garrity et al., 2002 ). However, since these rDNA regions are highly preserved areas, there is a limit to find polymorphism at level of inter-species such as subspecies, pathovar and race strains within species. Various molecular typing methods such as extragenic palindromic (REP) sequence, 124-127 enterobacterial repetitive intergenic consensus (ERIC) sequence 154 bp and BOX that are derived from repetitive sequences located in bacterial genomes have widely been used for discrimination of various bacterial strains at inter species level (De Brujin, 1992, Hulton et al., 1991) Recently, in silico genomic fingerprints were devised to be produced by virtual hybridization of 191 fully sequenced bacterial genomes using a set of 15,264 13-mer probes specially designed to produce universal whole genome fingerprints(Jaimes-Diaz et al., 2011).
Multiple arbitrary amplicon profile
(MAAP) that can universally be used in different organisms including pants, animals and microorganisms was developed for the use of single or multiple arbitrary primers (Caetano-Anolles, 1994; Caetano-Anolles and Gresshoff, 1997), and includes the techniques of RAPD (Williams et al., 1990),
AP-PCR (Welsh et al., 1990), and DNA amplification fingerprinting (DAF) (Caetano-Anolles et al., 1992). Because of its simplicity, the RAPD method has been used widely in studies of genetic diversity and in examining phylogenic and taxonomic relationship of bacterial strains (Caetano-Anolles and Gresshoff, 1997). However, in PCR techniques using arbitrary and RAPD primers, low reproducibility that causes variable PCR results depending on PCR condition has been recognized as a disadvantage factor (Caetano-Anolles et al., 1992).
A repeated DNA fragment (pKRD) of 1,187 bp was isolated from a genomic library of weedy rice distributed in Korea. A homology search showed the pKRD sequence belonged to a putative transposable element, the CACTA-like En/Spm family Furthermore, pKRD was used as a molecular marker to detect genetic variation of rice germplasm (Kang and Kang, 2008). Twenty primers consisting of 20 oligonucleotides were randomly designed from the repetitive
sequence, pKRD (Kang et al., 2002). Under the high stringent PCR condition, twenty primers of them can be applied in genomic fingerprinting of a variety of organisms including animals, other plants, and microorganisms, as well as rice and named as universal rice primer (URP). URP-PCR technology has been applied for accessing genetic diversity of various fungal and bacterial species. Numerous papers have demonstrated that URP-PCR profiles of fungal species are very useful for identifying microbial species at intra and inter species levels (Aggarwal et al., 2010. Aggarwal et al, 2010; Hong et al., 2008; Jana et al., 2005; Kang, 2012; Kang, 2001, Kang, 2002) and further the specific PCR polymorphic bands could be used for finding SCAR PCR primer for diagnosis of bacterial species or strain (Kang et al., 2003; Lim et al., 2007).
In this study, it was demonstrated that URP primers can universally be applied to PCR fingerprinting of diverse genomes from plant, human, animal and industry related bacterial strains at levels of intra and inter
species.
MATERIALS AND METHODS
Bacterial strains and genomic DNA extraction
Fourty one Agrobacterium vitis strains were isolated from different grapevein varieties by Hankyong National University. Twenty four strains of Pectobacterium carotovoum subsp. carotovorum, 3
Xanthomonas spp. 5 Pseudomonas spp, Rhizobium sp. of plant pathogenic bacteria were provided by Korea Agricultural Culture Collection (KACC). Themophilic bacterial strains containing Thermoactinomyces vulgaris that are randomly isolated form high temperature fermented compost were obtained from National Institute of Agricultural Sciences (NIAS), Rural Development Administration in Korea. DNA samples of human and animal related bacterial strains included 6 Escherichia coli strains (ATCC11105, 111a111b, M15, DH5, and O157:H7), 4 Salmonella spp., 7
Mycobacterium spp. and 3 Blucella abortus
strains were obtained from Medical School in Yeonsei University of Korea. Genomic DNA from gram negative and positive bacteria was extracted following the procedure described by Ausubel et al.(1987).
PCR analysis
URP with 20 mer were provided from JK BioTech, Co. Ltd., Ansung, Korea. PCR reactions were carried out in 50 ul reaction mixtures containing the DNA template (20 to 50 ng of purified DNA), 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 200 uM of each dNTP, 200 ng primer and 2.5 unit Taq DNA polymerase (Promega, USA). The reaction mixture was overlaid with a thin layer of sterile mineral oil to prevent evaporation. DNA amplification was performed in a programmable DNA thermal cycler (MJ Research, Inc., USA). The cycling parameters used were initial denaturation at 94oC for 4 min, followed by 35 cycles each consisting of 1 min at 94oC, 1 min at 55oC, and 2 min at 72oC. After the last cycle, the PCR tubes were incubated at 72C
for 7 min and were held at 4C. Amplified products were electrophoresed in a 1.5 % agarose gel in TAE buffer and visualized by staining with ethidium bromide.
PCR polymorphic bands based genetic relationship
PCR polymorphic bands were scored on their presence (value=1) or absence (value=0). The similarity coefficient was calculated by rearranging the scored bands of each rice variety. A dendrogram was constructed with the statistical program NTSYSpc (Rohlf, 2000) using the unweighted pair-group method with arithmetic mean (UPGMA).
RESULTS AND DISCUSSIONS
URP-PCR method was used to study the genetic diversity of 24 P. carotovurum subsp carotovorum (Pcc) strains causing bacterial soft rot disease on different plant species and 9 additive plant pathogenic bacteria. Fig. 1 shows PCR fingerprinting profile of diverse bacteria including
Pectobacterium carotovorum subsp.
carotovorum (Pcc), Xanthomonas
axonopodis pv. citri, Pseudomonas spp., Pseudomonas solanocearum, Rhizobium leguminosarum, Pseudomonas pv
phaseoliala, Pseudomonas marginalis,
Xanthomonas campestris pv. campestris and Pseudomonas syrinage pv syringae using URP 4. The PCR data revealed that the majority of the bacterial strains have multiple banding patterns showing their PCR polymorphisms. Furthermore, the results distinguished clearly difference among the Pcc strains, showing their high genetic diversity. In the case of Pseudomonas species, Pseudomonas pv phaseoliala and
Pseudomonas marginalis were divided as closely related group showing analogous PCR pattern among the Pseudomonas species. Agrobacterium vitis, a soilborn bacterium, is a causal agent of crown gall on grapevines. Crown gall disease of grapevine has rapidly become problematic in Korea because
tetraploid grapevine cultivars of Vitis vinifera (Kyohoand, Daebong), which are relatively sensitive to A. vitis, were intensively planted in farm fields (80%) of Ansung and Cheonan areas of Korea. Forty one strains of Agrobacterium vitis, the causal agent of crown-gall disease on grapevine (Fig. 2 A), originating from different geographical regions and 16 grapevine cultivars including 35 Kyoho cultivar of Korea, were characterized by PCR fingerprinting using URP 2 and URP 4(Fig. 2 B, C). A. vitis strains originated from Kyoho cultivar of grapevine showed relatively simple genetic diversity of the four PCR types, while the A. vitis strains originated from other grapevine cultivars showed various genetic diversities with 8 types. Comprehensively, UPGMA analysis based genetic relationship using the URP-PCR polymorphic bands showed 41 A. vitis strains are genetically clustered into large seven groups and 17 subgroups in the groups (Data not shown).
syringae
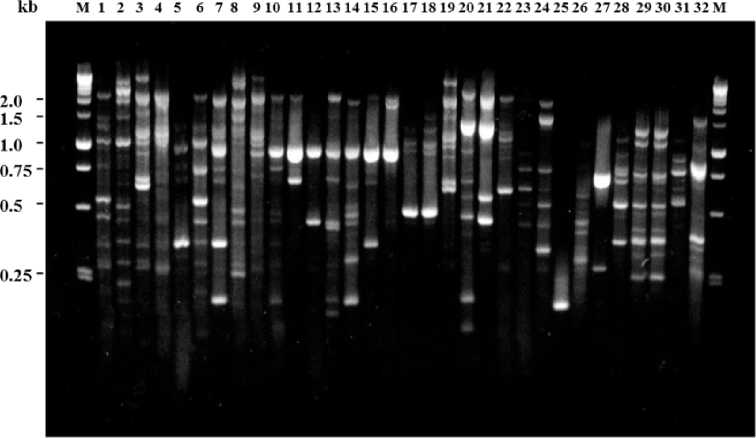
Fig. 1. PCR fingerprinting of different plant pathogenic bacterial strains by URP 4. M: 1kb DNA ladder (Promega), 1-24: Pectobacterium carotovoum subsp. carotovorum, 25: Xanthomonas axonopodis pv citri, 26: Pseudomonas sp. 27: Pseudomonas solanocearum, 28: Rhizobium leguminosarum, 29: Pseudomonase pv phaseoliala, 30: Pseudomonas marginalis, 31: Xanthomonas campestris pv. campestris, 32: Pseudomonas syrinage pv
Agrobacteriuin vitis strains
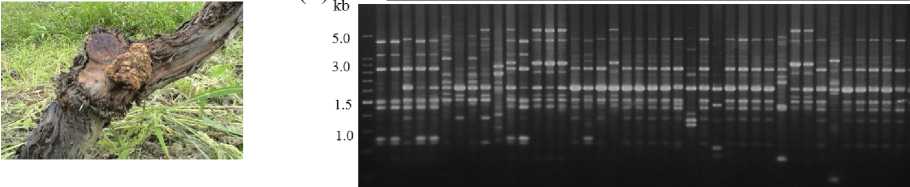
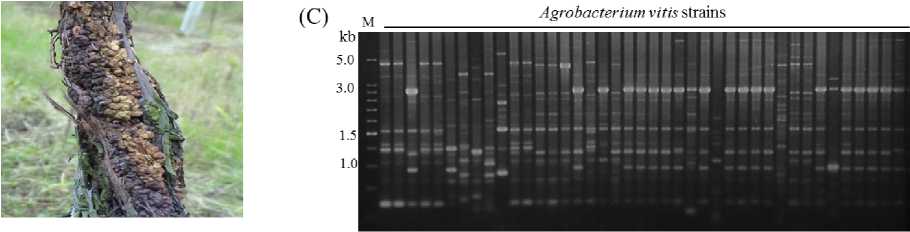
Fig. 2. URP-PCR polymorphism of Agrobacterium vitis strains, causal agent of crown gall on grapevine (A) detected by URP 2 (B) and URP 4 (C)
PCR-based methods are cheaper, easier and provide faster results; these methods include Random Amplified Polymorphic DNA PCR (RAPD-PCR), Repetitive extragenic palindromic PCR (REP-PCR) and Enterobacterial Repetitive Intergenic Consensus PCR (ERIC-PCR). These methods have been successfully applied for typing many bacteria (Anita et al. 2005; Johnson and Bryan, 2000; Williams et al. 1990). RAPD. Nevertheless, AP-PCR have been pointed out as low reproducibility problems as they are applied with annealing temperatures below 40oC. On the other hand, the URP-PCR can achieve stable PCR results because it has a high annealing temperature
of 55C. REP-PCR and ERIC-PCR fingerprinting methods using repeated sequence DNA of bacteria are limited in bacterial species retaining the sequence, whereas URP-PCR can universally apply in genomes from diverse bacteria strains without the reasonable sequence information on bacteria in advance. In particular, species or strain specific PCR fragment generated from URP-PCR products could be used in developing SCAR primers, which useful for specifically detecting Agrobacterium vitis and Pectobacterium carotovoum subsp. carotovorum (Kang et al., 2003; Lim et al., 2007).
Table 1. Oligonucleotide sequences of URP primers and their PCR applicability
on bacterial strains
|
Primers |
GC | |||
|
Sequences (5’-3’) |
Conte nt |
Tm (oC) |
Applicability on bacterial strains | |
|
URP1 |
ATCCAAGGTCCGAGACAACC |
50 |
65 |
Yes |
|
URP2 |
CCCAGCAACTGATCGCACAC |
50 |
65 |
Yes |
|
URP3 |
AGGACTCGATAACAGGCTCC |
50 |
66 |
Yes |
|
URP4 |
GGCAAGCTGGTGGGAGGTAC |
50 |
65 |
Yes |
|
URP5 |
ATGTGTGCGATCAGTTGCTG |
50 |
67 |
Yes |
|
URP6 |
TACATCGCAAGTGACACAGG |
50 |
68 |
No |
|
URP7 |
AATGTGGGCAAGCTGGTGGT |
55 |
74 |
Yes |
|
URP8 |
GATGTGTTCTTGGAGCCTGT |
50 |
65 |
Yes |
|
URP8 |
GGACAAGAAGAGGATGTGGA |
50 |
65 |
ND |
|
URP9 |
TACACGTCTCGATCTACAGG |
50 |
65 |
No |
|
URP10 |
AAGAGGCATTCTACCACCAC |
50 |
65 |
No |
Genomic DNA of 225 strains in 45 bacterial species were used as templates for URP-PCR
fingerprinting. The applicability of URP-PCR represents Yes, No and ND (Not determined)
In the following experiment, URP-PCR was applied to identification between or within human and animal -related bacterial species. As representative bacterial materials, genomic DNAs extracted from Mycobacterium spp. including 4 Mycobacterium intracellularae, strains, 4 M. avium strains, 3 M. scrofllaceum strains, M. fortuitum, M. gallinarum, M. canetti, and M. smegmatis. M. intracellularae, M. avium and
M. scrofllaceum strains that are the most common cause of cervical lymphadenitis in children, M. fortuitum is one of the many species of nontuberculosis mycobacteria (NTM) that are commonly found in the environment. M. canettii, a novel pathogenic taxon of the Mycobacterium tuberculosis complex (MTBC) (Hirsh et al., 2004), M. smegmatis is commonly used in work on the Mycobacterium genus due to its being a "fast
grower" and non-pathogenic (Simner et al., 2013).
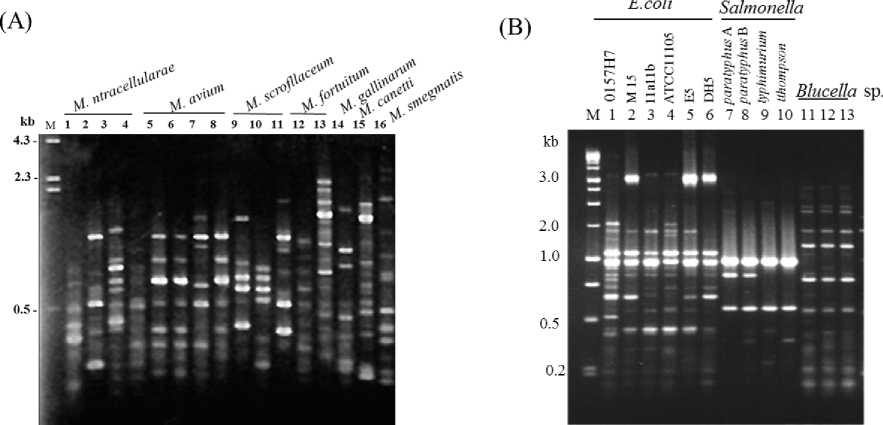
Fig. 3. PCR fingerprinting of human and animal related different bacterial species
Mycobacterium spp. (A), Echerichia coli strains, Salmonella spp. and Blucella (B) by URP2R.
Mycobacterium avium-intracellulare infection (MAI) is an atypical mycobacterial infection, i.e. one with nontuberculous mycobacteria or NTM, caused by Mycobacterium avium complex ("MAC"), which is made of three mycobacteria species, M. avium, M. intracellulare, and M. chimaera (Horsburgh.et al., 1985) This infection causes respiratory illness in birds, pigs, and humans, especially in immunocompromised people. As shown in Fig.3 (A), PCR polymorphism dependent on the strains was observed on PCR amplicons
of Mycobacterium spp., suggesting URP-PCR is useful for strain typing within the bacterial species. In addition, several human animal pathogenic bacteria were tested for confirming usefulness of URP-PCR. Non pathogenic Escherichia coli including strains ATCC11105, 111a111b, M15 and pathogenic E. coli O157:H7 that is one of the Shiga toxin-producing type and is a cause of disease, typically foodborne illness of the "colonic escherichiosis" type ( Furrer et al., 1990). E. coli strains, Salmonella spp. and Blucella abortus strains were used as
template DNA and were amplified by primer URP-3(Fig. 3B). The E. coli O157:H7 specific PCR polymorphic bands were observed on size marker of 0.5 kb and other polymorphic bands were showed in reliance on E. coli strains. As a result, six E. coli strains were divided into four types. Salmonella thompson, S. typhimurium, S. paratyphus A, and S. paratyphus B that are causal agents of Salmonellosis, food poisoning (McClelland et al., 2001; Bej et al., 1994), and animal and human pathogenic bacteria species such as Brucella strains were
also examined. A polymorphic band of 0.8 kb was observed on only S. paratyphus A, and S. paratyphus B, showing its specific PCR fragment in the bacterial species. Nevertheless, URP-PCR polymorphism of Blucella abortus strains was identical in the strains. Consequently, URP-PCR yielded distinct PCR profiles that permitted differentiation among them at interspecies level, and furthermore, ubiquitous strain types were observed within E. coli strains and Salmonella species.
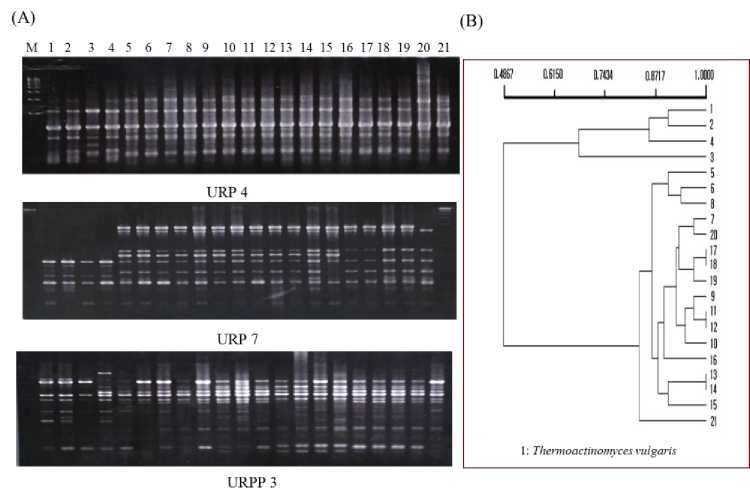
Fig. 4. URP-PCR profiles (A) and the PCR bands based UPGMA dendrogram (B)of
hemophilic bacterial strains isolated from compost fermented with high temperature
Finally, thermophilic bacteria that were randomly isolated from high temperature (45-55oC) fermented compost
was used for URP-PCR. Genomic DNAs were amplified by primers URP 3, URP 4 and URP 7 (Fig. 4 A) and resultant
polymorphic bands were scored to construct dendrogram. As showed in dendrogram data of Fig. 4 B, the bacteria were divided into two large groups containing culture collection, Thermoactinomyces vulgaris and further five subgroups were formed within the bacterial strains, showing different genetic relationships.
The data obtained from studies of various bacteria suggests that URP-PCR is potentially a valuable tool in the characterization and grouping at inter- and intraspecific levels of various bacterial species associated with medical, agricultural, industrial, and environmental fields. In many experiments using additional bacterial species, it was found URP-PCR allows generating discrete DNA fragments that are species or strain -specific. The speciesspecific URP-PCR products are easily excised from gels and cloned or re-amplified with the same primer to be used as a probe, providing an alternative strategy in developing specific Sequence-characterized amplified region (SCAR) markers that detect target microbial species or strain. Previously, such strategy was successfully used to develop specific DNA probe that detects Pectobacterium carotovorum subsp. carotovorum and A. vitis among other bacteria (Kang et al., 2003; Lim et al., 2006).
CONCLUSIONS
PCR approach using URP primers offers a powerful tool for studying DNA diversity of prokaryotic genomes, with potential use in taxonomic and phylogenic analysis, as well as in genotypic screening of strains in species, especially at inter- and intra-species levels. The URP-PCR is clearly different to RAPD (Williams et al., 1990) and AP-PCR method (Welsh et al., 1990) using arbitrary selected-primers because of long primer and high annealing temperature. It was believed that such URP-PCR conditions result in highly reproducible amplification products from organisms. This result suggested universal applicability of URP-PCR to human, animal, plant and industry related bacteria strains. Because URP primers can be applied to a wide range of bacterial species, mass screen of bacterial resources and database construction of DNA profile can be very useful for various purposes such as identifying genetic characteristics of specific strains. However, it remains to elucidate how URP primers designed from
repetitive sequence of rice are able to produce DNA polymorphism on diverse bacteria genomes with high specificity at inter- and intra-species. Nevertheless, it is reasonably assumed that complementary sequences to URP primers are widely dispersed in the diverse bacterial genomes that may play a critical evolutionary role.
REFERENCES
Ausubel, F. M. T., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., Steria ruhl, K. (1987). Current protocols in molecular biology. Greene Publishing Associates Wiley
Interscience, New York.
Aggarwal, R., Singh, V., Shukla, R., Gurjar, M., & Sharma, T. (2010). URP-based DNA fingerprinting of bipolaris sorokiniana isolates causing spot blotch of wheat. J. Phytopath, 158, 210-216.
Aggarwal, R., Tripathi, A., & Yadav, A. (2010). Pathogenic and genetic
variability in Tilletia indica monosporidial culture lines using universal rice primer-PCR Eur. J. Plant Pathol, 128, 333-342
Bej, A. K., Mahbubani, M. H., Boyce, M. J., & Atlas, R. M. (1994). Detection of Salmonella spp. In oysters by PCR. Appl. Environ. Microbial, 60, 368-373.
Caetano-Anolles, G. 1994. MAAP: a
versatile and universal tool for genome
analysis. Plant Molecular Biology, 25, 1011-1026.
Caetano-Anolles, G., & Gresshoff, P. M.
(1997). DNA markers: protocols, Applications, and overviews. WILEY-VCH, N.Y
Caetano-Anolles, G., Bassam, G. J., &
Gresshoff, P. M. (1992). Primertemplate interactions during DNA amplification fingerprinting with single arbitrary oligonucleotides. Mol. Gen. Genet, 235, 157- 165.
Chang, H. C., Wei, Y. F., Dijkshoorn, L., Vaneechoutte, M., Tang, C. T., et al. (2005). Species-level identification of isolates of the Acinetobacter calcoaceticus Acinetobacterbaumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J Clin Microbiol, 43, 1632–1639
Chang, H. R., Loo, L. H., Jeyaseelan, K., Earnest, L., & Stackebrandt, E. (1997). Phylogenetic Relationships of
Salmonella typhi and Salmonella typhimun'um Based on 16s rRNA Sequence Analysis. International Journal of Systematic Bacteriology, 47, 1253-1254.
De Brujin, F. J. (1992). Use of repetitive (repetitive extragenic pal-indromic and enterobacterial repetive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol, 58, 21802187.
Jaimes-Diaz, H., Garcia-Chequer, A. J.,
Mendez-Tenorio, A., Santiago-
Hemadez, J. C., Maldonado-Rodriguez, R., & Beattie, L. L. (2011). Bacterial classification using genomic
fingerprints obtained by virtual hybridization. J Microbiol Methods. 87, 286-94.
Furrer, B., Candrian, U., & Luethy, J. (1990). Detection and identification of
Escherichia coli producing heat-labile enterotoxin type I by enzymatic
amplification of a specific DNA
fragment. Lett. Appl. Microbial. 10, 3134.
Garrity, G. M., Johnson, K. L., Bell, J. A., & Searles, D. B. (2002). Taxonomic Outline of the Procaryotes. Bergey's Manual of Systematic Bacteriology, Second Edition, Release 3.0., SpringerVerlag, New York. 365 pages.
Hirsh, A. E., Tsolaki, A. G., DeRiemer, K., Feldman, M. W., & Small, P. M. (2004). Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci U S A, 101, 4871–4876.
Hong, S. K., Kim, W. K., Yun, H. G., & Choi, K. J. (2008).
Morphologicalvariations, genetic
diversity and pathogenicity
ofColletotrichum species causing grape ripe rot in Korea. Plant Pathol. J, 24, 269-278
Horsburgh, C. R, Mason, U. G., Farhi, D. C., & Iseman, M. D. (1985). Disseminated infection with Mycobacterium avium-intracellulare. A report of 13 cases and a review of the literature. Medicine, 64,
36-48.
Hulton, C. J. S., Higgins, C. F., & Sharp, P. M. (1991). ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typimurium and other enterobacteria. Molecular Microbiology, 5, 825-834.
Jana, T. K, Singh, N. K., Koundal, K. R., & Sharma, T. R. (2005). Genetic differentiation of charcoal rot pathogen, Macrophomina phaseolina, into specific groups using URP-PCR. Canadian J. Microbiol, 51, 159-164.
Johnson, J. R., & O’Bryan, T. T. (2000). Improved Repetitive-Element PCR fingerprinting for resolving pathogenic and nonpathogenic phylogenic groups within Escherichia coli. Clin Diagn Lab Immunol, 7:265-273
Kang, H. W. (2012). Genetic Divesity
Analysis of Fungal Species by
Universal Rice Primer (URP)-PCR.
The Korean Journal of Mycology, 40, 78-85.
Kang, H. W., Kwon, S. W., & Go, S. J. (2003). PCR-based specific and sensitive detection of Pectobacterium carotovorum ssp. carotovorum by primers generated from a URP-PCR fingerprinting-derived polymorphic band. Plant pathology, 52, 127-133.
Kang, H. W., Go, S. J., & Eun, M. Y. (2002). Fingerprinting of diverse genomes using PCR with universal rice primers generated from repetitive sequence of Korean weedy rice. Molecular Cells, 13, 1-7.
Kang, H. W., Park, D. S., Park, Y. J., You, C. H., Lee, B. M. and Go, S. J. (2001). Genomic differentiation among oyster mushroom cultivars released in Korea by URP-PCR fingerprinting.
Mycobiology, 29, 85-89.
Kang, H. W., Park, D. S., Park, Y. J., Lee, B. M., Cho, S. M., Kim, K. T., Seo, K. S., & Go, S. J. (2002). PCR based detection of Phellinus linteus using specific primers generated from universal rice primer (URP) derived PCR polymorphic band. Mycobiology, 30, 202-207.
Kang, H. W. &Kang, K. K. (2008). Genomic characterization of Oryza speciesspecific CACTA-like transposon element and its application for
genomic fingerprinting of rice varieties. Molecular Breeding, 21, 271-281.
Lim, S. H., Kim, J. K., & Kang, H. W. (2007). Novel SCARprimers forspecific and sensitive detection of Agrobacterium vitis strains. Microbiological Research, 164, 451-460.
McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., & Latreille, P. (2001). Complete genome sequence of Salmonella enterica serovar
Typhimurium LT2. Nature, 413, 852– 856.
Simner, P. J., Buckwalter, S. P., Uhl, J. R., & Wengenack, N. L. (2013).
Identification of Mycobacterium Species and Mycobacterium
tuberculosis Complex Resistance
Determinants by Use of PCR-
Electrospray Ionization Mass
Spectrometry. J Clin Microbiol, 51,
3492–3498.
Welsh, J., Petersen, C., & McClelland, C. (1990). Polymorphisms generated by arbitrarily primed PCR in the mouse: application to strain identification and genetic mapping. Nucleic Acids Res, 19: 303-306.
Williams, J. G. K., Kubelic, A. R., Livak, K. J., Rafalski, J. A., & Tingey, S. V. (1990) DNA polymorphisms amplified by arbitray primers are useful as genetic markers. Nucleic Acid Res, 18, 6531-6535.
64 • FACULTY OF AGRICULTURE, UDAYANA UNIVERSITY IN COOPERATION WITH
ASIA OCEANIA BIOSCIENCE AND BIOTECHNOLOGY CONSORTIUM
Discussion and feedback