MOLECULAR IDENTIFICATION OF EXOTIC FRUIT FLY Bactrocera occipitalis (DIPTERA: TEPHRITIDAE) USING MITOCHONDRIAL CYTOCHROME OXIDASE I (COI) GENE
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 6 No. 1 • September 2018
ISSN: 2303-3371
https://doi.org/10.24843/IJBB.2018.v06.i01.p04
MOLECULAR IDENTIFICATION OF EXOTIC FRUIT FLY Bactrocera occipitalis (DIPTERA: TEPHRITIDAE) USING MITOCHONDRIAL CYTOCHROME OXIDASE I (COI) GENE
I Putu Sudiarta1*, Dwi Martiningsia2, and I Nyoman Wijaya1 1Faculty of Agriculture Udayana University 2Agricultural Quarantine Agency Denpasar *Corresponding author: putu.ueda@yahoo.com
ABSTRACT
Some of fruit flies have been reported as the important pest on fruits and vegetables in the world. Agricultural Quarantine Agency Denpasar reported that there was new coming species (exotic) of fruit flies in Bali in 2014 based on the morphological identification, namely Bactrocera occipitalis. However Bactrocera dorsalis complex have similar morphological characters and have a less distinctive character for taxonomic identification, therefore it is difficult to identify fruit flies accurately. Based on that phenomena, the accurate identification is needed. One of the more accurate identification techniques is based on molecular identification using DNA-based barcode. To identify fruit flies, DNA-based barcode using mitochondrial cytochrome oxidase I (COI) gene has been conducted. PCR analysis using Fruit Fly MT-CO1-F (FFMT-CO1-F) 5’-GGAGCATTAATYGGRGAYG-3’ as forward primer and HCO 5’-TAAACTTCAGGGTGACCAAAAATCA-3’ as reverse primer was successfully amplified around 600 bp of COI gene of fruit flies. Based on similarity of sequence product, the species was identifiedas Bactrocera occipitalis and same result was revealed using morphological identification. Phylogenetic analysis of B. occipitalis based on COI genes showed that B. occipitalis from Bali were in the same groups with Bactrocera species from Tarakan and Philippines. In addition, Bactrocera occipitalis as exotic fruit fly is a new report in Bali, Indonesia.
Keywords: fruit flies, COI, Molecular identification, Phylogenetic analysis
INTRODUCTION
Fruit flies (Diptera: Tephritidae) are among the most destructive agricultural pests in the world, eating their way through acres and acres of citrus and other fruits at an alarming rate and forcing food and agriculture agencies to spend millions of dollars in control and management measures. Genus of Bactrocera is a group of fruit fly with 450 members of species (Drew and
Hancock, 2000). In addition some of spesies Bactrocera were reported as an importance pest on fruits and vegetables (Allwood et al., 1999). However, until now, the study of fruit flies has been traditionally biased towards applied aspects including the morphological identification, and lack of molecular approach. The traditional method for identification based on morphological characteristic has importance role to
understand species of fruit flies. On the other hand, the morphological identification has some weakness and not completely accurate. A morphological character occasionally is not describing the relationship with the genetic characters and some factors are caused by environmental interaction (Smith et al., 2003; Siwi, 2004). Based on those phenomena the more accurate identification is needed, one of them is molecular identification. Molecular identification has been utilized for improving the accurate information from morphological character, molecular characters more stable compared with morphological character (Hidayat, 2005). DNA barcoding has gradually been verified as an effective tool for identifying species in a wide range of taxonomic groups (Jiang et al., 2014). DNA barcoding using Gen Mitokondrial Cytochrome Oxidase Subunit I (MT-CO1) has been utilized for identification of fruit flies. The MT-COI barcode sequences for the diagnosis of fruit flies using 1426 sequences for 73 species of Bactrocera distributed worldwide was reported by Jiang et al. (2014). In addition, mitochondrial DNA also has been used to describe the genetic variation of animal (Loftus et al., 1994; Suryanto, 2003). As new information, Agricultural Quarantine Agency Denpasar reported that in 2013-2015 there was a new coming species or exotic fruit fly in Bali namely Bactrocera occipitalis based
on their morphological characteristics. In this study, DNA barcoding using MT-COI gene was performed to reconfirm both species.
MATERIALS AND METHODS
Fruit fly Trapp
The method of trapping of fruit fly was conducted base on protocol of survey of pests and diseases by Agricultural Quarantine Agency. Trapping area was designed all regencies around Bali using (Trapping) tipe Steiner (Steiner Trap) with Cue lure and Methyl eugenol attractant. The collecting of fruit fly was conducted every week to collect sample for morphological and molecular identification. Sample B. occipitalis Tarakan was collected from Laboratory of Agricultural Quarantine Agency Tarakan.
Morphological identification
The morphological identification was conducted as the first method to recognize the species of fruit fly. However, the method requires expert examination and may require additional supporting evidence such as the molecular diagnosis or host association records. Morfological identification was conducted based on identification key on book guide ”Fruit Flies of Indonesia : Their Identification, Pest Status and Pest Menegement” by Griffith University, Brisbane, Australia and The Australian Handbook for the Identification of Fruit Flies (Plant Health Australia, 2011). Key features
used for the morphological diagnosis of adult fruit flies include: wing morphology and infuscation, presence or absence of various setae, and relative setal size. (Note: Chaetotaxy, the practice, of setal taxonomy, is not as important in this group as some others.), overall colour and colour patterning, as well as presence, shape and color of thoracic vittae, a vitta is a band or stripe of colour (Plant Health Australia, 2011).
Molecular Identification
DNA extraction and MT-COI gen amplification
Qiagen DNeasy® (Qiagen, Hilden, Germany) Blood and Tissue kit was used to extract the DNA of fruit flies. The result of DNA extraction was amplified using PCR with general primers MT-CO1 Fruit Fly MT-CO1-F (FFMT-CO1-F)5’-GGA GCA TTA ATY GGR GAY G-3’ as forward primer and HCO 5’-TAA ACT TCA GGG TGA CCA AAA ATC A-3’ as reverse primer (Plant Health Australia, 2011). PCR amplification was performed in a 25 μl final reaction volume consisting of master mix 12,5 µl (1x BSA, 10x Buffer, dNTP’s, MgCl2, dan NEB Taq), 1 µl LCO1490-F, 1 µl HCO-R, DNA template 3 µl, dan Nuclease free water 7,5 µl., was conducted in Thermal cycler.The PCR thermal cycle program consisted of initial denaturation at 94 °C for 2 min, followed by 40 cycles of denaturation at 94
°C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 1 min with final extension at 72 °C for 7 min and finally holding at 4 °C until analyzed. (Plant Health Australia, 2011).
Electrophoresis, sequencing and phylogeny analysis
The PCR products with 1 µl loading dye were analyzed by gel electrophoresis on 2 % agarose gel at 70 V for 120 min in 1× TAE Buffer. DNA amplification product was visualized and captured by UV light using UV transilluminator at 302 nm wavelengths. The DNA target of both of fruit flies is around 600 bp (Delomen et al., 2013, Pramudi et al., 2013).
PCR product of fruit flies was analyzed in First Base Laboratorium, Malaysia for DNA sequencing., The sequence of nucleotide was used to search similarity in GeneBank with Program Basic Local Alignment Tool (BLAST) (NCBI 2014) on site of the European Bioinformatic Institute (EBI) (www.ebi.ac.uk). The homolog sequence of nucleotide of fruit flies was aligned to understand the similarity score using program ClustalW. Data from the sequencing of MT-COI DNA of fruit flies were analyzed using BioEdit 7.0.1 to determine the consensus sequences based on the conservative sequences and MEGA (Molecular Evolutionary Genetics Analysis)
6: 06 program to construct phylogeny trees (Tamura et al., 2013).
RESULTS AND DISCUSSION
The collected samples of fruit fly from trap were calculated in laboratory of Agricultural Quarantine Agency Denpasar.
After some treatments, all of the samples were identified. From the morphological key of new species or exotic species in Bali, the species was identified as Bactrocera occipitalis. Bactrocera occipitalis from Tarakan was use as control positive (Fig. 1).
Charateristic
B. occipitalis Bali B. occipitalis Tarakan
Caput

Thorax

Wing

Abdomen
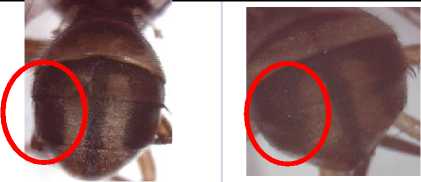
Fig. 1. morphological characteristic of b. Occipitalis. (a,b) Caput or head with large oval of spot on face, (c,d) black scutellum except of posterior and around prescutellar setae with brown colour, yellow stripe of scutum, (e,f) costal band overlapping R2+3, pointed of cubital streak (g,h) abdominal terga III-V orange-brown witha narrow to medium width medial longitudinal dark fuscous to black band over all three terga and lateral dark
DNA amplification
Mitocondrial cytochrome Oxidase I gene (MT-COI) of both fruit flies were successfully amplified by PCR using forward primer MT-CO1-F(FFMT-CO1-F)
5’-GGAGCATTAATYGGRGAYG-3’ and reverse primer: HCO 5’-
TAAACTTCAGGGTGACCAAAAATCA-3’, around 600 bp (Fig. 2). The PCR products were purified and used to sequence analysis.
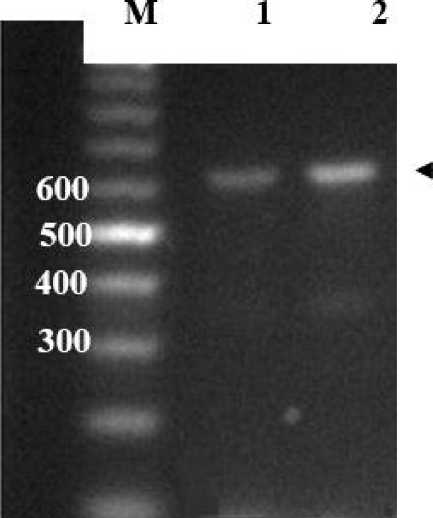
Fig. 2. PCR Product of amplification of MT-COI Gen of fruit flies. Lane M : marker 100 bp (Fermentas); 1. MT-CO1 gen of Bactrocera occipitalis Bali; 2. MT-CO1 gen of Bactrocera occipitalis Tarakan
Sequence homology 400
To understand the spe 300 and homology of fruit flies from Bali with other fruit flies, the nucleotide sequence of MT-CO1 gene was analyzed using software BLAST in Genebank. The homology
sequence of fruit fly with morphological character looks like B. occipitalis. The alignment data indicated MT-COI gen of B. occipitalis Bali has high similarity with MT-COI gen B.occipitalis from Filipina with homology 95,2 % (Table 1).
Table 1. The homology (%) sequence of of MT-COI gen of B. ocipitalis with other species Bactrocera in GenBank. [1] B. ocipitalis from Bali; [2] B. ocipitalis fromTarakan; [3] B. ocipitalis KC446122 Bd793 from Filipina; [4] B. ocipitalis KC446124 Bd 800 from Filipina; [5] B. ocipitalis KC446117 Bd 739 from Filipina; [6] B.
ocipitalis KC446107 Bd 783 from Filipina; [7] B. ocipitalis DQ116309 Isolat FF 1000 from i Filipina; [8] B. ocipitalis KC446110 Bd786 from Filipina; [9] B. ocipitalis KC446111 Bd787 from Filipina; [10] B. ocipitalis KC446105 Voucher Bd739 from Filipina; [11] Anasthrepa lugens HM538364 as outgroup; [12] B. caryae AY941232 as outgroup
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 12 | |
|
[1] |
ID | ||||||||||
|
[2] |
95 |
ID | |||||||||
|
[3] |
95 |
99,6 |
ID | ||||||||
|
[4] |
95,2 |
99,8 |
99,8 |
ID | |||||||
|
[5] |
95,2 |
99,8 |
99,4 |
99,6 |
ID | ||||||
|
[6] |
95 |
99,6 |
99,2 |
99,4 |
99,4 |
ID | |||||
|
[7] |
94,6 |
99,6 |
99,2 |
99,4 |
99,4 |
99,2 |
ID | ||||
|
[8] |
95 |
100 |
99,6 |
99,8 |
99,8 |
99,6 |
99,6 |
ID | |||
|
[9] |
948 |
99,8 |
99,4 |
99,6 |
99,6 |
99,8 |
99,4 |
99,8 |
ID | ||
|
[10] |
94,5 |
99,4 |
99 |
99,2 |
99,2 |
99,4 |
99 |
99,4 |
99,6 |
ID | |
|
[11] |
45,8 |
46,8 |
46,6 |
46,8 |
46,8 |
46,8 |
46,5 |
46,8 |
46,6 |
46,3 |
ID |
|
[12] |
37 |
37,4 |
37,4 |
37,4 |
37,4 |
37,4 |
37,2 |
37,4 |
37,4 |
37,4 |
34,3 ID |
Phylogeny tree and Genetic distance
Phylogeny analysis with unweighted pair group method (UPMGA) 100x bootstrap (Brinkman, F & D. Leipe 2001; Dharmayanti, 2011; Holil, 2004; Tamura et al., 2013) was used to construct phylogenetic tree of Bactrocera occipitalis Bali (Fig. 3).
Phylogenrtic tree showed that the B. occipitalis Bali is in the same clade with B. occipitalis Tarakan and B. occipitalis Philippines. The data also showed that B. occipitalis Bali is same monophyly group with B. occipitalis Tarakan and B. occipitalis Filipina (Fig. 3).
|
33 4" 69 99 100 |
β. occipitalis Tarakan DQ1163O9 B. occipitalis FF 100 Filipina KC446105 B. occipitalis Bd 798 Filipina KC446122 B. Occipitais Bd 793 Filipina KC446124 β. occpitais BdSOO Filipina KC446117 B. Occipitais Bd 739 Filipina KC446110 B. Occipitais Bd786 Filipina KC446107 B. Occipitais Bd783 Filipina | |
|
— |
KC446111 B. Occipitais Bd787 Filipina B. Occipitais BALI ⅝* ~ | <Aαas⅛ep⅛a Iudens B. Caiyae | |
|
2.0 |
Illl 1.5 1.0 0.5 0.0 | |
Fig. 3. Phylogeny tree of B. occipitalis and other Bactrocera from other countries base on
MT-CO1 gen
Genetic distance of MT-COI gen of B. occipitalis Bali with B. occipitalis Tarakan and Filipina around 0,043-0,052. The closer genetic distance of B. occipitalis Bali is with B. occipitalis KC446124 Voucher Bd800 Philippines and B. occipitalis KC446117 Voucher Bd793 Philippines which was 0,043 (Table 2).
The distribution of B. occipitalis is in The Philippines, Eastern Malaysia (Sabah), Brunei, as well as Indonesia (Kalimantan, West Java, dan Sumatara) (Drew and Romig, 2010). The main of host plants of B. occipitalis is Mangifera indica, Psidium
guajava, jeruk Citrus sp, Achras zapota and Averrhoa carambola (Clarke et al., 2005; Delomen et al., 2013; Yu et al., 2005; Allwood et al., 1999; Pujiastuti et al., 2009).
Table 2. Matrix of genetic distance of B. occipitalis Bali and other Bactrocera from other countries base on MT-CO1 gen. [1] B. occipitalis Bali [2]; B. occipitalis KC446117 Voucher Bd793 Filipina; [3] B. occipitalis KC446122 Filipina Voucher Bd 798; [4] B. occipitalis KC446124 Voucher Bd800 Filipina; [5] B. occipitalis KC446107 Vouher Bd783 Filipina; [6] B. occipitalis Tarakan; [7] B. occipitalis KC446105 Voucher Bd739 Filipina; [8] B.occipitalis DQ 116309 Filipina; [9] B. occipitalis KC446110; [10] B. occipitalis KC446111; [11] Anasthrepa lugens HM538364; [12] B. caryae AY941232
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 12 | |
|
[1] | |||||||||||
|
[2] |
0,043 | ||||||||||
|
[3] |
0,043 |
0,004 | |||||||||
|
[4] |
0,043 |
0,004 |
0,000 | ||||||||
|
[5] |
0,046 |
0,006 |
0,006 |
0,006 | |||||||
|
[6] |
0,046 |
0,002 |
0,002 |
0,002 |
0,004 | ||||||
|
[7] |
0,046 |
0,002 |
0,002 |
0,002 |
0,004 |
0,000 | |||||
|
[8] |
0,046 |
0,002 |
0,002 |
0,002 |
0,004 |
0,000 |
0,000 | ||||
|
[9] |
0,048 |
0,004 |
0,004 |
0,004 |
0,002 |
0,002 |
0,002 |
0,002 | |||
|
[10] |
0,052 |
0,008 |
0,008 |
0,008 |
0,006 |
0,006 |
0,006 |
0,006 |
0,004 | ||
|
[11] |
3,436 |
3,386 |
3,386 |
3,386 |
3,386 |
3,386 |
3,386 |
3,386 |
3,388 |
3,393 | |
|
[12] |
4,761 |
4,589 |
4,589 |
4,589 |
4,589 |
4,589 |
4,589 |
4,589 |
4,589 |
4,589 |
4,929 |
CONCLUSIONS
Based on morphological and molecular identification the Bactrocera occipitalis was found as a new coming species (exotic) of fruit fly in Bali.
ACKNOWLEDGEMENT
The current research was funded by collaborating between Magister of Agriculture Biotechnology, Faculty of
Agriculture Udayana University and Agricultural Quarantine Agency Denpasar.
REFERENCES
Allwood, A. J., Chinajariyawong, A., Drew, R. A. I., Hancock, D. L., Hengsawad, C., Jinapin, J. C., Jirasurat, M., Kong Krong, C., Leon, S., & Vijaysegaran, S. (1999). Kritsaneepaiboon, C.T.S. Host Plant records for fruit Flies (Diptera Tephritidae) in South-East Asia. Raffles Bullet. Zool. (Suppl.7).
Clarke, A. R., Armstrong, K. F., Carmichael, A. E., Milne, J. R., Raghu, S.,
Roderick, G. K., & Yeates, D. K. (2005). Invasive phytophagous pest
arising trough a recent tropical
evolutionary raditation: The
Bactrocera dorsalis complex of fruit flies. Quteprint.
Delomen, M. C. L., Mendioro, M. S., & Diaz, M. G. Q. (2013). Morphometric analysis and fruit flies Bactrocera occipitalis (Bezzi) and B. philipinensis Drew and Hancock (Diptera: Tephritidae) from Cavite and Davao del Norte. Philippine Journal of Science. 142 (1), 69-76.
Drew, R. A. I., & Romig, M. (2010). Fruit Flies. Biology, biosecurity, pest management and taxonomy.
International Centre for the Management of Pest Fruit Flies. Griffith University. Brisbane.
Drew, R. A., & Hancock, D. L. (2000). Phylogeny of the tribe Dacini (Dacinae) based on morphological, distributional, and biological data. In: Aluja M, Norrbom AL, editors. Fruit Flies (Tephritidae): Phylogeny and Evolution of Behavior. Florida: CRC Press, 491–504.
Hidayat, T. (2005). Systematic study of Subtribe Aeridinae (Orchidaceae) (PhD Thesis). Tokyo: The University of Tokyo.
Jiang, F., Jin, Q., Liang, L., Zhang, A. B., & Li, Z. H. (2014). Existences of
species complex largely reduced barcoding success for invasive
species of Tephritidae: a case study in Bactrocera spp. Molecular Ecology Resources. doi:10.1111/1755-
0998.12259.
Loftus R. T., MacHugh D. E., Bradley D. G., Sharp P. M. & Cunningham, P. (1994). Evidence for two independent domestications of cattle. Proceedings of the National Academy of Sciences
of the United States of America 91, 2757–61.
Martiningsia D., Wijaya, I. N., & Sudiarta, I. P. (2017). Karakteristik Molekuler Dan Filogeni Lalat Buah Bactrocera occipitalis (Diptera: Tephritidae) dari Tarakan Berdasarkan Sekuen Nukleotida Gen Coi. J. Agric. Sci. and Biotechnol, 6(1).
Plant Health Australia. (2011). The Australian Handbook for the Identification of Fruit Flies. Version 1.0. Plant Health Australia. Canberra, ACT.
Pramudi, M. I., Puspitarini, R. D., &
Rahardjo, B.T. (2013).
Keanekaragaman dan kekerabatan lalat buah (Diptera:Tephritidae) di Kalimantan Selatan berdasarkan
karakter morfologi dan molekuler (RAPD PCR dan Sekuensing DNA). J HPT Tropikal, 13(2), 191 – 202.
Pujiastuti, Y., Kartini, L., Irsan, C., Herlinda, S., Adam, T., & Thalib, R. (2009). Variasi jenis tanaman buah sebagai inang lalat buah (Diptera:Tephritidae) Pada berbagai ketinggian tempat di Sumatra Selatan. Jurnal Agripet Fakultas Pertanian Palangka Raya Kalimantan Tengah, 10(2), 58-64.
Siwi, S. S. (2004). Jenis-jenis lalat buah penting di Indonesia dan macam tanaman inangnya. Bogor: Balai
Besar Penelitian dan Pengembangan Bioteknologi dan Sumber Daya
Genetik Pertanian.
Smith, P. T., Kambhampati, S., & Amstrong, K. A. (2003). Phylogenetic relationships among Bactrocera spesies (Diptera : Tephritidae)
Inferred from mitochondrial DNA sequences. Manhattan: Molecular Phylogenetics and Evolution, 8-17.
Suryanto, D. (2003). Melihat
keanekaragaman organisme melalui beberapa teknik genetika molekuler. Medan: Program studi biologi
fakultas matematika dan ilmu
pengetahuan alam universitas sumatera utara.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular Evolutionary
Genetics Analysis Version 6.0. Mol. Biol. Evol. 30(12), 2725–2729.
Yu, D. J., Chen, Z. L., Zhang, R. J., & Yin, W. Y. (2005). Real time qualitative PCR for the Inspection And identification Of Bactrocera
philipinensis And Bactrocera occipitalis (Diptera:Tephritidae)
using Sybr Green Assay. The Raffles Buletin of Zoology, 53(1), 73-78.
FACULTY OF AGRICULTURE, UDAYANA UNIVERSITY IN COOPERATION WITH
ASIA OCEANIA BIOSCIENCE AND BIOTECHNOLOGY CONSORTIUM • 43
Discussion and feedback