DETECTION METALLO-BETA-LACTAMASE GENE IMP-1 AND IMP-2 OF Pseudomonas aeruginosa CLINICAL ISOLATES IN SANGLAH HOSPITAL BALI
on
DETECTION METALLO-BETA-LACTAMASE GENE IMP-1 AND IMP-2 OF Pseudomonas aeruginosa CLINICAL ISOLATES IN SANGLAH HOSPITAL BALI
Ni Made Adi Tarini1,2*, Ni Nengah Dwi Fatmawati1,2,3, and I Putu Bayu Mayura1 1Department of Clinical Microbiology, Medical School, Faculty of Medicine, Udayana University 2Clinical Microbiology Laboratory, Sanglah General Hospital 3Molecular Biology Laboratory, Faculty of Medicine, Udayana University *Corresponding author : nmatarini@unud.ac.id
ABSTRACT
Pseudomonas aeruginosa is a pathogen frequently found as an agent of Hospital Acquired infections. This bacterium is very easy to be resistant to several types of antibiotics through various mechanisms. Carbapenem such as Imipenem and Meropenem is a potential option for the therapy of this bacterium, but unfortunately P. aeruginosa has ability in hydrolyzing these antibiotics through enzyme metallo-β-lactamases (MBLs). Recently, IMP and VIM, MBLs enzyme group are reported common from various countries, but no data is reported for these enzymes in Indonesia especially in Bali. In fact, the resistant data of P. aeruginosa against carbapenem group antibiotics such as meropenem and imipenem is quite high in Sanglah General Hospital in 2014 was 35% and 45% respectively. Therefore, the aim of this study was to detect IMP-1 and IMP-2 genes of MDR P. aeruginosa, which are phenotypically resistant to the antibiotic Imipenem and Meropenem disks based on CLSI standards in Clinical Microbiology Laboratory, Sanglah General Hospital, Denpasar, Bali. Eighty-six isolates were isolated from sputum (25 / 29.1%), wound (25 / 29.1%), urine (15 / 17.4%), endotracheal Tube (11 / 12.8), pus (6/7% ), blood (3 / 3.5%) and tissue (1 / 1.1%). In this study, all isolates were subjected to PCR for detection of IMP-1 and IMP-2. The result showed that 9 isolates were positive IMP-1 gene (10.5%), but there was no isolate positive for IMP-2 gene. The result was similar with that of the other countries, especially for the gene IMP-1. Detection and molecular characterization of MBL-producing P. aeruginosa strains are very important for infection control purposes. Currently, this study is still continued for detection of another MBL genes.
Keyword : Pseudomonas aeruginosa, MBLs gene, PCR, Positive Control
INTRODUCTION
Bacterial Multidrug Resistance (MDR) is an indicator of the changes in the strains of bacteria that are often associated with the administration of antibiotic unwisely, such as unnecessary empirical therapy, the selection of antibiotic regimens improperly, and prolonged use of antibiotics. One of bacteria found in hospital setting that easy to become an MDR bacteria is Pseudomonas aeruginosa. It is a pathogen frequently found as an agent of Hospital Acquired infections. This bacterium is very easy to be resistant to several types of antibiotics through various mechanisms. Prolonged use of antibiotics causing this bacteria to become resistant immediately
against several groups of antibiotics such as Beta-lactam, Aminoglycosides, Chloramphenicol, Quinolone, Tetracycline and Sulphonamides. Thereby making the infections caused by these bacteria very difficult to treat ( Arunagiri K et al.,2012). Carbapenem, such as Imipenem and Meropenem, is a potential option for the therapy of this bacterium, but unfortunately P. aeruginosa has an ability in hydrolyzing these antibiotics through enzyme metallo-β-lactamases (MBLs). (Zhao WH et al., 2009). Prevalence rates of P. aeruginosa produce MBLs vary widely. Study in India in 2012 reported MDR P. aeruginosa isolate produced MBLs in 70.1% of isolates, which was higher than that in 2002 that is only
12% (Arunagiri K et al., 2012). Several types of MBLs reported from Enterobacteriaceae and Gram-negative nonfermenter group bacteria such as IMP, VIM, SPM, SIM and GIM isolated from clinical specimens. Enzymes such as IMP and VIM are two groups of enzymes were predominantly found in the previous studies (Giske CG et al., 2006). Moreover, both of enzymes are also found predominantly in Asia. Recently, IMP and VIM, MBLs enzyme group are reported commonly found from various countries. Study in India, 2012, the MDR P. aeruginosa isolates showed that both of MBLs gene such as blaVIM genes and blaIMP gene were 87.2% and 4.3%, respectively. However, it has been different with study conducted in Iran in the same year, which reported blaVIM gene and blaIPM gene were18.2% and 9,6%, respectively. (Sepehriseresht S et al., 2012). No data is reported for MBLs enzymes in Indonesia especially in Bali. In fact, the resistant data of P. aeruginosa against Carbapenem group antibiotics such as Meropenem and Imipenem is quite high in Sanglah General Hospital in 2014 was 35% and 45% respectively. Based on this condition, therefore, the aim of this study was to detect IMP-1 and IMP-2 genes of MDR P. aeruginosa, which are
phenotypically resistant to the antibiotic Imipenem and Meropenem disks based on CLSI standards in Clinical Microbiology Laboratory, Sanglah General Hospital, Denpasar, Bali using PCR.
MATERIALS AND METHODS
Bacterial isolates and identification
Eighty-six glycerol stock isolates of P. aeruginosa were cultured on MacConkey
Agar and incubated aerobically at 35±2°C, 18-24h. The isolates were isolated from sputum (25 / 29.1%), wound (25 / 29.1%),
urine (15 / 17.4%), Endotracheal Tube (11 / 12.8), pus (6/7%), blood (3 / 3.5%) and tissue (1 / 1.1%) during 2013-2015.
Identification of P. aeruginosa and
drug susceptibility test by VITEK-2 based on CLSI Standard.
Bacterial Genomic DNA Isolation
Isolates of P. aeruginosa that have grown on MacConkey agar plates were harvested around 5-10 colonies and suspended in 200 μl PBS (Phosphate Buffered Saline) pH 7.3. Bacterial genomic DNA was isolated by using Roche High Pure PCR Template Isolation Kit (Roche Life Science, Indianapolis, USA) based on manufacturer’s instruction from bacterial suspension. DNA was eluted with 50 μl of elution buffer.
PCR for blaIMP1 and blaIMP2
gene
PCR was conducted using Go Taq® Green Master Mix (Promega, Madison, USA). Uniplex PCR to detect 16S rRNA gene was performed. It was followed by Uniplex PCR to detect IMP-1 and
IMP-2 gene used primer Listed in Table 1. The final concentration of primers were 0.4 μM each. Protocols of PCR were described in previous study (Sepehriseresht et al, 2012). PCR cycle was initiated with pre denaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 1 min, annealing primer at 55°C (16sRNA and IMP-1) and 48oC (IMP-2) for 1 min and extension at 72°C for 1 min; and final extension at 72°C for 7 min (iCycler, Biorad thermal cycler).
ISSN ONLINE: 9 772303 337 008
Table 1. Primer used for Gene VIM and IPM Detection (Shibata et al.,2003)
|
No. |
Nama Primer Sekuens Primer (5'÷ 3’) bp |
|
1. |
blaIMP1 Forward: ACC GCA GCA GAG TCT TTG CC 587 Reverse: ACA ACC AGT TTT GCC TTA CC |
|
2. |
blaIMP2 Forward: GTT TTA TGT GTA TGC TTC C 678 Reverse: AGC CTG TTC CCA TGT AC |
Amplicons were electrophoresed on 1.5% agarose gel in TBE buffer at 60 volt, and for 35 min. DNA was visualized with GelRed™ Nucleic Acid Gel Stain (Biotium, Hayward, CA 94545) and
RESULTS AND DISCUSSION
A total of 86 glycerol stock isolates of P. aeruginosa from clinical samples, which were phenotypicall resistant to antibiotics imipenem and meropenem using VITEK-2 based on CLSI Standard, were suscessfully cultured on MacConkey agar media plates. The colonies showed pale yellow colonies. All clinical isolates were positive tested for the presence of 16sRNA
gene (Fig.1), confirmed that all isolates were P. aeruginosa. All isolates were subjected to PCR for detection of IMP-1 and IMP-2. The result showed that 9 (10.5%) isolates were positive IMP-1 gene (Fig.2), but none of isolate was positive for IMP-2 gene. The study to detect MBLs gene have been conducted in the other countries. Study in Iran showed that 6.6% and 3.3% of the P. aeruginosa had IMP-1 and IMP-2 genes, respectively (Sepehriseresht S et al,2012). In Asian Countries and regions, two MBLs gene are commonly prevalent. Study in Japan showed IMP-1 gene was 0.5% and IMP-2 was 64.5% (Shibata et al, 2003), whereas in China reported that IMP-1 type of MBLs was 89.7% (Arunagiri K, 2012)
500 bp
100 bp
1 2 3 4 5
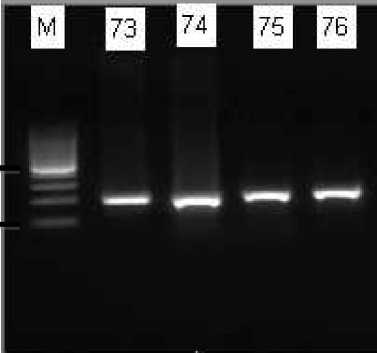
214 bp
Fig. 1. PCR for 16sRNA gene detection. The expected band was 214 bp. Amplicon was electrophoresed on 1.5% agarose gel. (M = Marker 100 bp; Lane 2 - 5 = 16sRNA positive band = 214 bp)
1 2 3 4 5 6 7 8 9 10
500 bp
100 bp
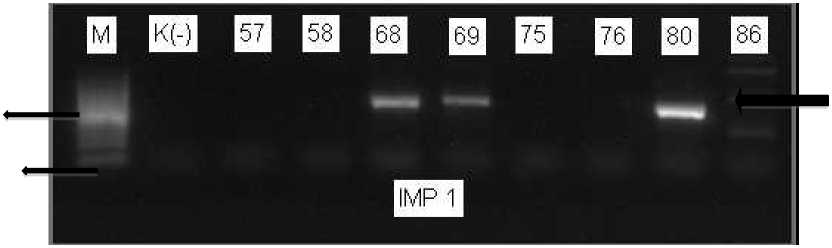
587 bp
Fig. 2. PCR for IMP-1 gene detection. Lanes 5,6 and 9 were positif for IMP-1 gene (587 bp). Amplicon was electrophoresed on 1.5% agarose gel. (lane 1 = marker, lane 2 = negative control )
This study showed the similar condition with the MBL gene research studies from the other countries, especially for the gene IMP-1. However, it was slightly different for IMP-2 gene. Detection and molecular characterization of MBL-producing P. aeruginosa strains very important for infection control purposes. Currently, this study is still continued for detection of another MBL genes.
ACKNOWLEDGMENTS
This work was financially supported by Hibah LITBANG Medicine Faculty Udayana University, Bali, Indonesia under Grant No. No. 2672/UN14/KU/2014 . We
thank Wahyu Hidayati (Molecular Biology Laboratory staff), Putu Yuliandari, M.D. (Clinical Microbiology Laboratory, Faculty of Medicine staff), and Ni Wayan Nilawati (Clinical Microbiology Laboratory Sanglah General Hospital staff) for their technical supports.
REFERENCES
Arunagiri, K., Sekar, B., Sangeetha, G., John, J. (2012). Detection and Characterization of Metallo- β-Lactamases in Pseudomonas
aeruginosa by phenotypic and
Molecular Methods from
Clinical Samples in
Tertiary Care Hospital, West Indian Med J, 61(8), 778-783.
Bush, K., Jacoby, G.,A., Medeiros, A.,A. (1995). Afunctional classification scheme for beta lactamases
and its correlation with molecular structure. Antimicrob Agents Chemother., 39, 1211-1233.
Castanheira, M., Toleman, M.,A., Jones, R.,N., Schmidt, F.,J., Walsh,T.,R. (2004). Molecular
characterization of a β-lactamase gene, blaGIM-1,
encoding a new subclass of metallo-β lactamase. Antimicrob
Agents Chemother., 48, 4654-4661.
Giske, G.,C., Libisch, B., Colinon, C., Scoulica, e., et al. (2006). Establishing Clonal Relationships between VIM-1-like Metallo-β-Lactamases-Producing Pseudomonas aeruginosa Strain from Four European
Countries by Multilocus Sequence Typing, J. Clin. Microbiol, 44(12), 4309-4315.
Heinz, U., Adolph, H.,E. (2004). Metallo-β-lactamases: two binding sites for one catalic metal ion. Cell Mol Life
Sci, 61, 2827-2839.
Hanson, N.,D., Hossain, A,, Buck, L., Moland, E.,S., Thomson, K.,S.
ISSN ONLINE: 9 772303 337 008
(2006) First occurrence of a Pseudomonas aeruginosa isolate in the United States producing an IMP metallo-β-lactamase, IMP-18.
Antimicrob Agents Chemother., 50, 2272-2273.
Lauretti, L., Riccio, M.,L., Mazzariol, A., et al. (1999) Cloning and
characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother, 43, 1584-1590.
Mahon, C.,R. (2011). Textbook of
Diagnostic Microbiology.
Poirel, L., Naas, T., Nicolas, D. (2000). Characterization of VIM-2, a
Carbapenem Hydrolyzing
Metallo-β-Lactamase and its Plasmid and Integron-Borne Gene from a Pseudomonas aeruginosa Clinical Isolate in France. Antimicrob Agents Chemother., 44, 891-897.
Shibata, N., Doi, Y., Yamane, K., et al.(2003). PCR Typing of Genetic determinats for metallo-β-l
actamases and integrases carried by gram-negative bacteria isolated in Japan with focus on the class 3 integron. J Clin Microbiol., 41, 5407-5413.
Sepehriseresht, S., Boroumand, M.,A., Pourgholi, L., et al. (2012). Detection of VIM and IPM-type
Metallo-β-Lactamases in
Pseudomonas aeruginosa Clinical Isolates. Archives of Iranian
Medicine, 15(11), 670-673.
Sacha, P., Wieezorek, P., Hauschild, T., Zorawski, M., et al. (2008). Metallo-β-Lactamases of Pseudomonas
aeruginosa – a Novel Mechanism Resistance to β-Lactam Antibiotics. Folia Histochemica Et
Cytobiologica, 46(2), 137-142.
Toleman, M.,A., Bennett, P.,M., Walsh, T.,R. (2006) ISCR Elements: novel gene-capturing systems of the 21st century?, Microbiol MolBiol Rev., 70, 296-316.
Woodford, N., Turton, J., F., Livermore, D.,M. (2011). Multiresistant Gramnegative Bacteria: the Role of High-Risk Clones in the Dissemination of Antibiotic Resistance, FEMS Microbiol Rev, 35, 736-755.
Yan, J.,J., Hsueh, P.,R., Ko, W.,C., et al. (2001). Metallo-β-Lactamases in
clinical Pseudomonas isolates in
Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents
Chemother., 45, 2224-2228.
Zhao, W.,H., Chen, G., Ito, R., Qing Hu, Z. (2009). Relevance of Resistance Levels to Carbapenems and
Integron-Borne blaIMP-1, blaIMP-7, blaIMP-10, and blaVIM-2 in Clinical Isolates of Pseudomonas
aeruginosa, Journal of Medical Microbiology, 58, 1080-1085.
36 • ASIA OCEANIA BIOSCIENCES AND BIOTECHNOLOGY CONSORTIUM
Discussion and feedback