SUGAR PRODUCTION BY DIGESTING OF OIL PALM EMPTY FRUIT BUNCH USING EXTRACELLULAR ENZYMES FROM Aspergillus niger AND Trichoderma reesei FOR ETHANOL PRODUCTION
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • VOL. II NO. 1 • SEPTEMBER 2014
ISSN: 9 772303 337008
SUGAR PRODUCTION BY DIGESTING OF OIL PALM EMPTY FRUIT BUNCH USING EXTRACELLULAR ENZYMES FROM Aspergillus niger AND Trichoderma reesei FOR ETHANOL PRODUCTION
Kahar Muzakhar1*, Sutoyo1, Siswoyo2
1Biology Dept., Faculty of Math. and Nat. Sci., The University of Jember, Indonesia. 2Chemistry Dept., Faculty of Math. and Nat. Sci., The University of Jember, Indonesia. *Corresponding author: kaharmzk@unej.ac.id
ABSTRACT
Extracellular enzymes which obtained from 4 days cultivation Aspergillus niger and Trichoderma reesei on solid state fermentation of oil palm empty fruit bunch (OPEFB) were used for lignocellulosic--‐‑rich OPEFB digestion. The enzymes were concentrated using 70% saturated ammonium sulphate, dialysed against 20mM acetate buffer at pH 5 and adjusted one tenth (v/v) from the initial volume with the same buffer The concentrated enzymes were then used in hydrolysation of powdered OPEFB. Amount of 10.65 mg/ ml and 11.47 mg/ml sugars were produced when each concetrated enzyme A. niger and T. reesei mixed with2%OPEFB. These hydrolysation were done on 100 ml total volume, incubated at 37oC with 100 rpm shaken for 36 hours. Further, both hydrolyzates results were sterilised and fermented anaerobically using Saccharomycess cerevisiae at concentration 0.5mg/ml cells and incubated in 30oC for 24 hours. Colorimetric analysis using QuantiChrom Kit DIET--‐‑500 at OD 580nm gave results the alcohol production were 0.86% and 0.92% which were similar with Gas Chromatograph analysis that of 0.83% and 0.93%, respectively.
Keywords: extracellular enzymes, hydrolysation, fermentation
INTRODUCTION
Itisforecasted,inyear2000–2020Indonesia is the
largest producer and exporter of oil palm in the international market (Sumathi
et al., 2008). A huge amount of lignocellulosic material oil palm empty fruit bunch (OPEFB) is generated during production of oil palm from the fruit and remained as agriculture waste product (Baharuddin et al., 2013; Huzairi et al., 2013) Abundant of OPEFB is accumulated in the field, and considered to be difficult materials to digest (Purwandari et al., 2013) which consists of three main polymeric components, i.e., cellulose (46.7%), hemicellulose (17.9%),and lignin (4.2%) (Quintero et al., 2011). Thus, OPEFB is a common waste problem in oil palm plantations Unadventurously, they are burned resulting in air pollution and massive residues on landfills. To solve the disadvantages, several of processing methods i.e., biologically (Huzairi et al., 2013; Zhang et al., 2013) mechanical (Baharuddin et al., 2013; Shamsudin et al., 2011), chemical (Kim and Ho 2013; Rahman et al., 2007) have been investigated which shift the structural and chemical compositions of OPEFB lignocellulose
to sugars (Ye et al., 2014) and was converted to renewable energy bioethanol (Cui et al., 2014 Hoon et al., 2011). Further, researchs related to OPEFB utilization, have also been developed to produce biogas (Mohamed et al., 2013; O--‐‑thong et al., 2013; She et al., 2013), enzymes (Ariffin et al., 2008; Ottenheim et al., 2014), sugar and oligosaccharides derivates (Ling A et al., 2014 Ottenheim et al., 2014; Rahman et al., 2006) and other products. However, in fact, little attention is put forward on OPEFB utilisation in terms of economic point of view. In this paper, microbial utilization of OPEFB to produce sugar using crude extract from A.niger and T. reesei, and conversion of its hydrolysates to ethanol will be reported.
MATERIALS AND METHODS
Extracellular Enzymes Production
Source extracellular enzymes were obtained from solid culture of one kilogram sterilized OPEFB in a ten liter flask which each culture was inoculated with A. niger and T. reesei. The enzymes were harvested after optimum incubation at 30oC for 5 days. Enzymes extraction
was done by adding 1000 ml water containing 1% NaCl, shaken at room temperature for 9 hours and filtered using paper filter on funnel Buchner To remove remaining cells from the filtrate, centrifugation at 4000 rpm for 20 minutes was performed. The supernatant which containing enzymes were then concentrated by ammonium sulfate at 70% saturation, dissolved on 30 ml acetate buffer pH5. The remaining ammonium sulphate was removed through dialysis on mPES MicroKros Filter Modules C02--‐‑E010--‐‑05--‐‑S 10 KDa against the same buffer for 24 hours, and then enzyme solution brought to one tenth (v/v) from the initial volume using buffer above. The two concentrated enzymes were stored at 4oC until needed for OPEFB hydrolysis.
Enzymatic Hydrolysis OPEFB
OPEFB hydrolysis was done by using concentrated extracellular enzymes containing 2% powdered OPEFB. Two grams of powdered OPEFB was suspended into 100 ml concentrated crude enzyme, incubated at 37oC, shaken 100 rpm, 36 hours. Enzyme reaction was stopped by boiling at 100oC for 10 minutes. The resulting supernatant as hydrolysate was recovered and separated from remaining OPEFB substrate by filtration on paper filter and then centrifuged at 4000 rpm for 10 minutes. Hydrolysate was stored at 4oC for next sugar analysis. This experiment was carried out twice.
Sugar Analysis of OPEFB Hydrolysate
Total sugar of OPEFB hydrolysate was estimated by phenol--‐‑sulfuric colorimetric method (Dubois, et al., 1956). The reducing sugar of hydrolysate was also measured using Somogy (Somogyi et al., 1926) and Nelson (Nelson et al., 1944) methods. Sugar components of hydrolysate were analysed by Gas Chromatograph (GC) Thermo Scientific Trace 1310 equipped withTG--‐‑ 225MS 15m x 0.25mm x 0.25µμm column. GC was setup at 190°C (5 minute hold) to 250°C at 8°C/ min (5 minute hold) and carrier gas hydrogen adjusted at flow rate 45cm/sec. One milliliter OPEFB hydrolysate sample was transmethylated
as alditol acetate (Arai and Murao 1978; Spiro et al., 1972). An amount 0.5µμl of sample was injected to GC for analysis. The hydrolysate was also analyzed by using TLC in silica gel plate (Merck, 60 F254). A mixture (v/v) of butanol : ethanol : chloroform : amonia (4 : 7.5 : 4 : 8) was used as solvent system and 0.1% sulfuric acid containing 0.1% vanilin was employed for detection.
Anaerobic Fermentation and Analysis of Ethanol Concentration
In anaerobic fermentation to produce ethanol, yeast cells S. cerevisiae was used. The yeast was pre--‐‑cultured aerobically in 100 ml medium pH 6 in 1 liter shaker flasks, containing yeast--‐‑extract (0.3%), malt extract (0.3%), pepton (0.5%) and glucose (1%). The culture was incubated at 25°C under shaker set at 120 rpm for 24 hours. Active yeast cells were harvested by centrifugation at 4000 rpm for 5 minutes. The pellet was suspended into sterilized water with the concentration of cells 25 mg cells/ ml and further used for source inoculum in anaerobic fermentation of OPEFB hydrolysate Fermentation was done in 50 ml medium using mini fermenter at 30oC, inoculated with 25 mg cells (≈1ml) of source yeast prepared above. The ethanol concentration was analysed using QuantiChrom Kit DIET--‐‑500 colorimetric method at OD 580nm. Ethanol concentration was also analysed using GC above equipped with Trace GOLD TG--‐‑1301MS GC column with sample volume 0.5µμl. The GC machine was setup at 250°C with carrier gas helium at flow rate 35cm/sec. The remaining sugar component after fermentantion was analysed with the same method as described earlier at Sugar Analysis of OPEFB Hydrolysate.
RESULTS AND DISCUSSION
Optimization and Production of Extracellular Enzymes
Extracellular enzyme produced by A. niger and T. reesei in OPEFB medium were done without any nutrient added that has been confirmed. For
preliminary step, the optimization of enzyme extracellularproductionfromthetwospecieswere done in small scale using 10 gr OPEFB medium Evidence revealed that in 3 days cultivation, A. niger and T. reesei grow well in OPEFB medium Much mycelium with their black spores covered whole of OPEFB medium. Additionally, OPEFB was also contain crude protein nearly 0.3% on dry--‐‑basis which was needed as a nitrogen source for fermentation. Mean, A. niger and T. reesei certainly utilized carbon and nitrogen from OPEFB for their growth. Further, we observed that in 5 days cultivation, small amount liquid phase was produced, indicating hydrolysation process happen. Accordingly, some extracellular enzymes were released by both A. niger either T. reesei during solid state fermentation. The extracellular enzyme was then harvested by 1% NaCl extraction. The enzyme activity against 1% OPEFB alkali extract substrate was measured daily based on reducing sugar produced. Thus, incubation days of cultivation to produce extracellular enzyme released optimally can be represented as maximum of enzyme activity to produce reducing sugar against OPEFB alkali extract substrate. The activity levels of extracellular enzymes were, therefore, determined for incubation periods of 1 to 7 days as shown at Figure 1.
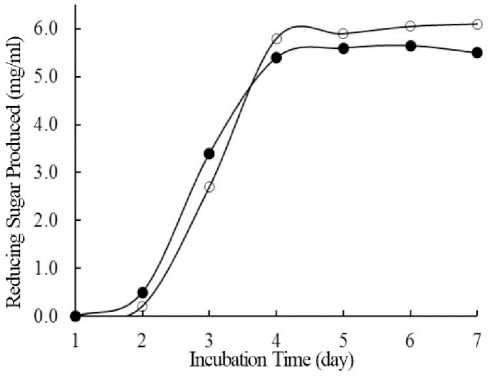
Fig 1. Optimizing of extracellular enzymes production of A. niger (®) and T. reesei (S) on OPEFB.
As stated in Figure 1, extracellular enzymes of A. niger and T. reesei has highest activity to hydrolysed OPEFB alkali extract substrate when incubation time was done in 4 days. Both isolates produced reducing sugars 5.4 mg/ml and 5.8 mg/ml, respectively. Based on this result, the enzyme production was scaled up in 1 kg of OPEFB medium. The enzymes activity were also examined and gave similar results compare with previous small scale enzymes production, where the enzymes hydrolysed OPEFB alkali extract and produced 5.1 mg/ml and 5.3 mg/ml reducing sugars. It was confirmed already that genus Aspergillus (Alam et al., 2011; Ottenheim et al., 2014) and Trichoderma (Shafawati et al., 2013; Wang Z et al., 2014) release some extracellular enzymes which decomposed cellulose, hemicellulose, and lignocellulose of OPEFB.
OPEFB Hydrolysis and Sugar Analysis
Hydrolysis was carried out in 100 ml concentrated enzymes containing 2% of powdered OPEFB and incubated at 37oC with 100 rpm shaken for 36 hours. The hydrolysis process by the two enzymes were measured every 4 hours. They produced optimum of reducing sugar production in between 32 and 36 hours incubation. The longer incubation time, the more higher the degree of hydrolysis. And thereafter 40 hours the hydrolysis is still in progress but not significant increasing in reducing sugar production and could presumably attain similar degree of hydrolysis at very much later time Analysis revealed that enzymes from A. niger and T. reesei released 10.65 mg/ml and 11.47 mg/ ml reducing sugars after 36 hours hydrolysis. In addition, OPEFB consist of 66.5% total sugar (or 13.3 mg/ml when the suspension contain 2% OPEFB)measuredbyphenol--‐‑sulfuriccolorimetric method (20)so that it can be concluded that concentrated enzyme from A. niger and T. reesei digested OPEFB reach 81.7% and 89.2% degree of hydrolysis, respectively. To improve the degree of hydrolysis, optimization of hydrolysation must be done, e.g., pH, temperature and period of incubation time. In another words to make
the process more effective, suitable pretreatment and enzymatic reaction parameters need to be optimized (Hassan et al., 2013).
As shown at Table 1, phenol sulfuric acid analysis showed that the total sugar of OPEFB hydrolysates were 10.86 mg/ml and 11.87 mg/ ml respectively when hydrolysis of OPEFB were done by concentrated enzymes from A. niger and T. reesei. However, GC analysis revealed that OPEFB hydrolysates contain much sugar as monosaccharides, that of10.65 mg/ml and 11.47 mg/ml. Means, OPEFB which hydrolysed by both concentrated enzymes from A.niger and T. reesei produced sugar rich hydrolysates as monosaccharides but poor in oligosaccharides The monosaccharides concentration in hydrolysates were 98.1% and 96.6%. TLC analysis displayed that the hydrolysis product was primarily monosaccharide glucose spot respectively (figure not shown). The ability of both extracellular enzymes from A. niger and T. reesei to release monosaccharides from complex polysaccharides due to that species could produce broad spectrum of extracellular enzymes. The capability of crude enzyme to hydrolyze OPEFB and produce glucose and xylose as monosaccharide could be due to the fact that some genus Aspergillus and Trichoderma produce of various enzymes such as cellusases, glucoamylase and xylanase (Chen et al., 2014; Chandra et al., 2009; Krijgsheld et al., 2013; Perrone et al., 2007).
Analysis of Anaerobic Fermentation
Fermentation to produce ethanol was done using OPEFB hydrolysate as medium without any adjusting of either pH or sugar concentration Further, both hydrolyzates results were sterilised and fermented anaerobically using S. cerevisiae at concentration 0.5mg/ml cells and incubated in 30oC for 24 hours. Colorimetric analysis using QuantiChrom Kit DIET--‐‑500 at OD 580nm gave results the alcohol production were 0.86% and 0.92% which were similar with Gas Chromatograph analysis that of 0.83% and 0.93% The remaining sugar in hydrolysate after fermentation as monosaccharides was also analysed by GC. After 24 hours fermentation, glucose concentration were very low with concentration value 0.12 mg/ml and 0.18 mg/ml But, the xylose almost nearly same comparing with initial concentration, that of 0.53 mg/ml and 0.71 mg/ml, correspondingly. This evidence demonstrated that S. cerevisiae difficult ferments monosaccharide xylose from OPEFB hydrolysate, similar result as reported by Sudiyani et. al. 2013 But in contrast S. cerevisiae definitely utilized and converted glucose to ethanol. By these results, improvement hydrolysis OPEFB must be done which expected to increase the yield of sugar as monosachharides in OPEFB. Adjusting and optimizing environment factors in hydrolysis and as well as parameters in fermentation such as sugar concentration, pH of medium and temperature are also needed (Ferreira et al., 2009; Shill et al., 2012; Viell et al., 2013).
Tabel 1. Sugar Analysis of OPEFB Hydrolysates
Sugar concentration (mg/ml) in OPEFB
hydrolysis using concentrated enzyme A. niger Hydrolysate analysis
(A) and T. reesei (B)
A
Gas Chromatograph Analysis (Monosaccharides component as alditol acetates)
|
--‐‑ Glucose |
8.81 |
8.93 |
|
--‐‑ Xylose |
1.11 |
2.12 |
|
--‐‑ Others |
0.73 |
0.42 |
|
Total monosaccharides |
10.65 |
11.47 |
|
Phenol Sulfuric Acid Analysis | ||
|
Total Sugar |
10.86 |
11.87 |
CONCLUSION
In this study, bioconversion of potential cheap material of OPEFB to ethanol was successfully done. And it is necessary to develop more efficient method with the target to improve the ethanol yield as well as for profitability reason process. Enzymatic hydrolysis and fermentation process must be evaluated in order obtain high ethanol yield Xylose as pentose sugar has not been consumed by S. cereviseae. Therefore, assessment of other pentose--‐‑consuming microorganisms with the aim to completely ferment the sugars released in OPEFB hydrolysates must be investigated.
ACKNOWLEDGEMENTS
This work was supported by Directorate General of Higher Education (DIKTI) Ministry of National Education and Culture --‐‑ Indonesia through competitive research funding scheme Riset Strategis Nasional 2014.
REFERENCES
Alam MZ, Bari MN, Muyibi SA and Jamal P 2011 Development of Culture Inoculum for Scale--‐‑Up Production of Citric Acid from Oil Palm Empty Fruit Bunches by Aspergillus niger. Procedia Environ. Sci. 8, November, pp. 396–402.
Arai M and Murao S. 1978. Characterization of Oligosaccharides from an Enzymatic Hydrolyzate of Red Yeast Cell Walls by Lytic Enzymes. Agric. Biol. Chem. 42, 9: 1651–1659.
Ariffin H, Hassan MA, Shah UK, Abdullah N, Ghazali FM and Shirai Y. 2008 Production of Bacterial Endoglucanase from Pretreated Oil Palm Empty Fruit Bunch by Bacillus pumilus EB3. J. Biosci. Bioeng. 106, 3: 231– 236.
Baharuddin AS, Sulaiman A, Kim DH, Mokhtar MN, Hassan MA, Wakisaka M, Shirai Y and Nishida H. 2013. Selective component
degradation of oil palm empty fruit bunches (OPEFB) using high--‐‑pressure steam. Biomass and Bioenergy. 55: 268–275.
Chandra M, Kalra A, Sangwan N S, Gaurav S S, Darokar MP and Sangwan R S. 2009 Development of a mutant of Trichoderma citrinoviride for enhanced production of cellulases. Bioresour. Technol. 100, 4: 1659– 62.
Chen X, Luo Y, Yu H, Sun Y, Wu H, Song S, Hu S and Dong Z. 2014. Transcriptional profiling of biomass degradation--‐‑related genes during Trichoderma reesei growth on different carbon sources. J. Biotechnol. 173: 59–64.
Cui X, Zhao X, Zeng J, Kheang S, May Y and Liu D. 2014. Bioresource Technology Robust enzymatic hydrolysis of Formiline--‐‑pretreated oil palm empty fruit bunches (EFB) for efficient conversion of polysaccharide to sugars and ethanol. Bioresour. Technol. 166: 584–591.
Dubois M, Gilles KA, Hamilton JK, Rebers AP and Smith F. 1956. Colorimetric Method for Determination of Sugars and Related Substances,” Anal. Chem. 28, 3: 350–356.
Ferreira S, Duarte AP, Ribeiro JA, Queiroz, and Domingues F. C. 2009. Response surface optimization of enzymatic hydrolysis of Cistus ladanifer and Cytisus striatus for bioethanol production,” Biochem. Eng. J. 45, 3: 192–200.
Hassan O, Pei T, Yusof M and Muhammad N 2013. Optimization of pretreatments for the hydrolysis of oil palm empty fruit bunch fiber (EFBF) using enzyme mixtures.
Biomass and Bioenergy. 56, 0: 137–146
Hoon Y, Jung I, Han J, Choi I and Heon K.
2011. Bioresource Technology Aqueous ammonia pretreatment of oil palm empty fruit bunches for ethanol production.
Bioresour. Technol. 102, 20: 9806–9809.
Huzairi M, Zainudin M, Ali M, Tokura M and Shirai Y. 2013. Bioresource Technology Indigenouscellulolyticandhemicellulolytic bacteria enhanced rapid co--‐‑composting of
lignocellulose oil palm empty fruit bunch with palm oil mill effluent anaerobic sludge. Bioresour. Technol. 147: 632–635.
Kim S and Ho C. 2013. Bioethanol production using the sequential acid alkali--‐‑pretreated empty palm fruit bunch fi ber. Renew. Energy 54: 150–155.
Krijgsheld P, Bleichrodt R, Van--‐‑Veluw GJ, Wang F, Müller W H, Dijksterhuis J, and Wösten H aB. 2013. Development in Aspergillus. Stud. Mycol. 74, 1: 1–29.
Ling A, Carvalheiro FL, Duarte C, Roseiro L B, Charalampopoulos D and Rastall R A 2014. Bioresource Technology Production and purification of xylooligosaccharides from oil palm empty fruit bunch fibre by a non--‐‑isothermal process. Bioresour. Technol. 152: 526–529.
Mohamed P, Jahim J, Harun SM, Markom O, Hassan and Wahab A. 2013. Biohydrogen production from pentose--‐‑rich oil palm empty fruit bunch molasses: A first trial,” Int. J. Hydrogen Energy. 38, 35: 15693– 15699.
Nelson N. 1944. A Photometric Adaptation Of The Somogyi Method For The Determination Of Glucose,” J. Biol. Chem. 153: 375–380.
Ottenheim C, Verdejo C, Zimmermann W, and Wu JC. 2014. Hemicellulase production by Aspergillus niger DSM 26641 in hydrothermal palm oil empty fruit bunch hydrolysate and transcriptome analysis. J. Biosci. Bioeng. xx: 1–6.
O--‐‑thong S, Boe K and Angelidaki I. 2012 Thermophilic anaerobic co--‐‑digestion of oil palm empty fruit bunches with palm oil mill effluent for efficient biogas production. Appl. Energy. 93: 648–654.
Perrone G, Susca A, Cozzi G, Ehrlich K, Varga J, Frisvad JC, Meijer M, Noonim P, Mahakarnchanakul W and Samson RA 2007. Biodiversity of Aspergillus species in some important agricultural products. Stud. Mycol. 59: 53–66.
Purwandari FA, Sanjaya AP, Millati R, Cahyanto M N, Horváth IS, Niklasson C and
Taherzadeh MJ. 2013. Pretreatment of oil palm empty fruit bunch (OPEFB) by N--‐‑ methylmorpholine--‐‑N--‐‑oxide (NMMO) for biogas production: structural changes and digestion improvement. Bioresour. Technol. 128: 461–6, Jan.
Quintero A, Cardona A and Piarpuza D. 2011 Empty fruit bunches from oil palm as a potential raw material for fuel ethanol production. 5: 3–10.
Rahman SHA, Choudhury JP, Ahmad AL, and Kamaruddin AH. 2007. Optimization studies on acid hydrolysis of oil palm empty fruit bunch fiber for production of xylose. 98: 554–559.
Rahman SHA, Choudhury JP and Ahmad AL 2006. Production of xylose from oil palm empty fruit bunch fiber using sulfuric acid. 30: 97–103.
Shafawati SN and Siddiquee S. 2013. Composting of oil palm fibres and Trichoderma spp. as the biological control agent: A review.
Int. Biodeterior. Biodegradation. 85: 243–253, Nov.
Shamsudin S, Kalsom U, Zainudin H, and Abd--‐‑ aziz S. 2011. Effect of steam pretreatment on oil palm empty fruit bunch for the production of sugars. Biomass and Bioenergy. 36: 280–288.
She P, Jahim J and Harun S. 2013. Enhancement of batch biohydrogen production from prehydrolysate of acid treated oil palm empty fruit bunch. Int. J. Hydrogen Energy 38, 22: 9592–9599.
Shill K, Miller K, Clark DS and Blanch HW 2012. A model for optimizing the enzymatic hydrolysis of ionic liquid--‐‑ pretreated lignocellulose. Bioresour. Technol. 126: 290–7.
SomogyiM.1926.NotesOnSugarDetermination,” J. Biol. Chem. 70: 599–612.
Spiro RG. 1972. Methods in Enzymology Study of the carbohydrates of glycoproteins Analysis of Monosaccharides. Acad. Press. New York London Vol.XXVIII. Ginsbg. I.V., ed.), Part B. 28.
Sudiyani Y, Styarini D, and Triwahyuni E. 2013 Utilization of biomass waste empty fruit bunch fiber of palm oil for bioethanol production using pilot – scale unit. Phys. Procedia. 32: 31–38.
Sumathi S, Chai SP and Mohamed AR. 2008 Utilization of oil palm as a source of renewable energy in Malaysia, Renew. Sustain. Energy Rev. 12 (9): 2404–2421.
Viell J, Wulfhorst H, Schmidt T, Commandeur U, Fischer R, Spiess A and Marquardt W. 2013 An efficient process for the saccharification of wood chips by combined ionic liquid pretreatment and enzymatic hydrolysis. Bioresour. Technol. 146: 144–51.
Wang Z, Bay H, Chew K and Geng A. 2014 High--‐‑loading oil palm empty fruit bunch saccharification using cellulases from Trichoderma koningii MF6,” Process Biochem. 49, 4: 673–680.
Ye L, Sufian M, Hudari B, Li Z and Chuan J 2014. Simultaneous detoxification, saccharification and co--‐‑fermentation of oil palm empty fruit bunch hydrolysate for l --‐‑lactic acid production by Bacillus coagulans JI12. Biochem. Eng. J. 83: 16–21.
Zhang Y, Sun W, Wang H and Geng
A. 2013. Bioresource Technology Polyhydroxybutyrate production from oil palm empty fruit bunch using Bacillus megaterium R11. Bioresour. Technol. 147: 307–314.
38 • asia oceania biosciences and biotechnology consortium
Discussion and feedback