Comparison of DNA Yield from Different Plant Materials of Plumeria sp. (Apocynaceae)
on
Journal of Advances in Tropical Biodiversity and Environmental Sciences 3(1): 8-11, February 2019
(p-ISSN: 2549-6980)
DOI: 10.24843/atbes.v03.i01.p03 Available online at: https://ojs.unud.ac.id/index.php/ATBES/article/view/48682
8
Comparison of DNA Yield from Different Plant Materials of Plumeria sp. (Apocynaceae)
Vanesa Martida and Made Pharmawati*
Biology Study Program, Faculty of Mathematics and Natural Sciences, Udayana University, Kampus Bukit
Jimbaran, Badung, Bali
*Corresponding author: made_pharmawati@unud.ac.id
Abstract. DNA extraction that gives good quantity and quality DNA is a basic step that must be completed for molecular studies, especially in DNA fingerprint imaging. The aim of this research was to find out the better quality and quantity of DNA extracted from different plant materials of frangipani cultivars (Plumeria sp.). Leaves and flowers were collected from Taman Jepun, Denpasar Bali. Fresh young leaves and flowers were used as plant materials as well as dried leaves (silica gel dried leaves) of Plumeria sp. This research used CTAB buffer with modification as lysis buffer. Purification techique used NucleoSpin® Gel and PCR Clean Up Kit. The results showed that the colour of DNA solution from fresh material was clear and the quantities of DNA from young fresh leaves were between 70-300 ng/µl. The DNA colour solution from flowers was also transparent with concentration between 0-40 ng/µl. DNA isolated from dry material resulted in brown solution with DNA quantity between 30100 ng/µl and need to be purified to obtain clear DNA solution.
Keyword: DNA extraction, leaves, flower, Plumeria sp.
I. INTRODUCTION1
Plumeria is a plant genus in Apocynaceae family. This plant is commonly used as garden plant. Plants in this family are known to have various secondary metabolites such as alkaloid and cardenolid that are used as traditional medicine to relieve fever, asthma and whooping cough [1].
There are high numbers of Plumeria sp and cultivars known, and they need to be characterised for inventory of germplasm. Characterisation can be done using molecular markers. Various molecular marker techniques can be used to analysed their genetic characters [2]. Analysis using molecular marker requires the availability of high quality DNA.
There are various methods of plant DNA extraction, from self-prepare extraction buffers to commercial DNA extraction kits. Commercial DNA extractions kits have been reported to have several advantages such as shorten extraction time and resulted in high purity DNA [3]. However, it also has limitation due to high cost.
Among many methods using self-prepared buffer, extraction using CTAB buffer has been mostly used. It has been used for extraction of DNA from various plants including woody plants [4], medicinal plants [5], and aquatic plants [6]. The CTAB method used detergent based buffer that contain cetyl trimethylammonium bromide to purify DNA from plant tissue [7].
The phenolic compound in plant tissue often caused difficulties in DNA extraction [8]. The phenolic compound that present in DNA solution can inhibit enzymatic reaction such as polymerase chain reaction (PCR). Modification of extraction protocols and DNA purification need to be done to proceed the enzymatic reaction [9].
Source of tissue for DNA extraction has also influence the success of DNA extraction. Commonly, DNA was extracted from fresh young leaves or frozen young leaves. DNA can also be extracted from dry samples although force drying can degrade DNA [10]. The aim of this research was to compare plant materials used for DNA extraction of Plumeria sp. .
II. RESEARCH METHODS
Plant Materials
Plant materials used were fresh young leaf and flower of Plumeria alba ‘Moonlight’, P. alba ‘Cendana HB#3’, P. rubra ‘Cheddi Pink’, P acuminata ‘Sudamala Bali’, and P. acuminata ‘Sudamala Sassy’. Leaf and flower samples were collected from Taman Jepun, Denpasar, Bali.
Silica gel dried plant materials were prepared by collecting young leaves of P. acuminata ‘Sudamala Bali’, P. acuminata ‘Bali Mas’, P.acuminata ‘Maroon’, P. alba ‘Bali Hai Gold’, P. alba ‘Bali Palace’, P. rubra ‘Cheddi Pink’, P. Obtusa, Plumeria sp. ‘Jack Purple’ and Plumeria sp. ‘Madam Poni’. Leaf samples were put into plastic bag
DOI: 10.24843/atbes.v03.i01.p03 Available online at: https://ojs.unud.ac.id/index.php/ATBES/article/view/48682
9
containing silica gel. The silica gels were replaced everyday.
solution became clear (Fig 3). Visualization of purified DNA using agarose gel electrophoresis was shown in Fig. 4.
DNA extraction
DNA extraction was done according to Doyle dan Doyle [7] with modification. The concentration of EDTA in the lysis buffer of this study was higher than that of Doyle and Doyle [7]. The lysis buffer contained 2% w/v CTAB, 1.4 M NaCl, 50 mM EDTA, 100 mM Tris-HCL (pH 8), 0,2% (v/v) 2-mercaptoethanol. The size of leaf and petal flower used was 1.5 cm x 1.5 cm (without main vein for leaves samples). The materials were ground using mortar and pestle and 1 ml lysis buffer was added.
The mixture was incubated at 65ºC for 30 minutes. After that, 1x volume of chloroform: isoamilalcohol (24:1) was added, vortexed and centrifuged at 8.000 rpm for 10 minutes. The aquous layer was transferred to a new tube and 2/3x volume of cold isopropanol was added. The mixture was incubated overnight at -20oC.
Following incubation, centrifugation was done for 5 minutes at 8.000 rpm. Pellet DNA was washed with 500 µl of 70% ethanol and the centrifugation was repeated for 5 minutes. Pellet DNA was air dried and then the pellet was dissolved in 100 µl sterile aquadest.
DNA Purification
DNA purification was done on DNA extracted from silica gel dried leaves which had brown colour. Purification was conducted using NucleoSpin® Gel and PCR Clean Up Kit (Macherey-Nagel) according to company instruction.
DNA Electrophoresis
Electrophoresis was done using 1% agarose gel in 1x TAE buffer (40 mM Tris, 20 mM acetic acid and 1 mM EDTA) stained with Ethidium bromide [11]. As much as 3 µL DNA was loaded to the gel and the electrophoresis was run at 100 volt for 30 minutes. The concentration of DNA was estimated using lambda DNA with known concentrations. DNA was visualized using UV transiluminator.
III. RESULTS AND DISCUSSION
Results
DNA extracted from fresh leaves and flowers of Plumeria sp. using modified CTAB method resulted in clear DNA solution. Result of DNA electrophoresis was shown in Fig. 1. The concentrations of DNA from fresh young leaves were between 70-300 ng/µl, while the DNA concentrations from fresh flowers were between 0-40 ng/µl.
DNA extraction from silica gel dried leaves resulted in DNA solution with brown colour (Fig. 2a). Visualiation of DNA extracted from silica dried Plumeria sp leaves is shown in Fig 2b. The estimated DNA concentrations ranged from 30 ng/µl to 100 ng/µl.
DNA purification was done for DNA extracted from silica gel dried leaves. Following purification, the DNA
1 2 3 4 5 6 7 8 9 10 λ1 λ2
High MW
DNA
Smeared
DNA ]
X.
RNA -£
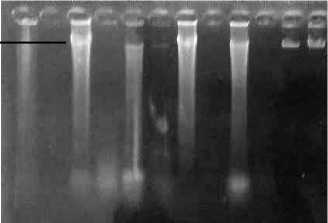
Fig. 1. Electrophoresis of DNA extracted from fresh leaves and flower of Plumeria sp. 1, 3, 5, 7, 9 were from fresh leaves, 2,4,6,8,10 were from fresh flower. 1-2 P. alba ‘Moonlight’; 3-4 P. alba ‘Cendana HB#3’; 5-6 P. rubra ‘Cheddi Pink’; 7-8 P. acuminata ‘Sudamala Bali’; 9-10 P. acuminata ‘Sudamala Sassy’; λ1= lambda DNA 200 ng; λ2= lambda DNA 400 ng
a
b

12 34 56789
1 2 3 4 5 6 7 8 9 100bp
High MW DNA
RNA
Smeared DNA
Fig 2. Brown colour of DNA solution extracted from silica gel dried leaves (a) and agarose gel electrophoresis of DNA from dried leaves samples (b) 1: Plumeria acuminata ‘Sudamala Bali’; 2: P. acuminata ‘Bali Mas’; 3: P.
acuminata ‘Maroon’; 4: P. alba ‘Bali Hai Gold’; 5: P. alba ‘Bali Palace’; 6: P. rubra ‘Cheddi Pink’; 7: P. obtusa; 8:
Plumeria sp. ‘Jack Purple’; 9i: Plumeria sp. ‘Madam Poni’
DOI: 10.24843/atbes.v03.i01.p03 Available online at: https://ojs.unud.ac.id/index.php/ATBES/article/view/48682
10

Fig. 3. Clear DNA solution from dried leaves after purification. 1: Plumeria acuminata ‘Sudamala Bali’; 2: P. acuminata ‘Bali Mas’; 3: P. acuminata ‘Maroon’; 4: P. alba ‘Bali Hai Gold’; 5: P. alba ‘Bali Palace’; 6: P. rubra ‘Cheddi Pink’; 7: P. obtusa; 8: Plumeria sp. ‘Jack Purple’; 9i: Plumeria sp. ‘Madam Poni’,
1 2 3 4 5 6 7 8 9 λ1 λ2
High MW DNA
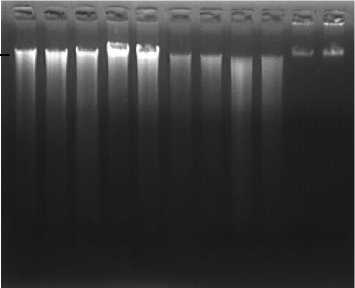
Fig. 4. Agarose gel electrophoresis of DNA from dried leaves after purification 1: Plumeria acuminata ‘Sudamala
Bali’; 2: P. acuminata ‘Bali Mas’; 3: P. acuminata ‘Maroon’; 4: P. alba ‘Bali Hai Gold’; 5: P. alba ‘Bali
Palace’; 6: P. rubra ‘Cheddi Pink’; 7: P. obtusa; 8: Plumeria sp. ‘Jack Purple’; 9: Plumeria sp. ‘Madam Poni’, ; λ1= lambda DNA 150 ng; λ2= lambda DNA 250 ng
Discussion
Examination on the quantity and quality of DNA was conducted using agarose gel electrophoresis. The range of concentration of genomic DNA extracted from fresh leaves of Plumeria sp. was 70 ng/µl to 300 ng/µl. The concentration of DNA obtained from fresh flower was between 0 – 40 ng/µl. High concentration of DNA in fresh leaf could be due to different weight of leaf and flower used. The weight of 1,5 cm x 1,5 cm leaf differed from 1,5 cm x 1,5 cm flower petal. Leaf of Plumeria sp is thicker than flower petal. Besides that, flower of Plumeria sp contains essential oil such as geraniol, farsenol, sitronelol and linalool [12]. These essential oils can inhibit the process of DNA extraction.
Gel electrophoresis of DNA showed smeared pattern which indicated that the DNA has degraded. The extraction of DNA did not use liquid nitrogen during tissue grinding. The grinding of tissue can be made easier with liquid
nitrogen, because it freezes the cell and avoid DNA degradation [13].
Drying samples in silica gel has an advantage for collection of sample in remote areas, where samples cannot quickly transfer to laboratory for further process. Silica gel absorbs moisture and liquid particles easily from the air chamber and helps to resist tissue damage. Another advantage of silica gel is that it is an inert substance (difficult to react) with surrounding substances, so it does not damage existing tissue sample [14]. It was reported that high quality of DNA was obtained from common bean leaf tissue preserved in silica gel [15]. However, in this research, DNA extracted from silica dried Plumeria leaves had brown colour with concentration of 30 ng/µl to 100 ng/µl. The brown colour of DNA is caused by phenol oxidation [8, 16]. Generally, phenolic compound is found in meristem tissue, such as in young leaf [13]. Phenolic compound can reduce yield of DNA and reduce DNA purity. Oxidized polyphenols bind to DNA and precipitate with DNA resulting in a brown color [17]. In this study, the CTAB method is more appropriate for extracting DNA from fresh leaf.
To purify the brown colour DNA, NucleoSpin® Gel dan PCR Clean Up Kit was used and resulted in clear DNA solution. During DNA purification, several contaminants such as secondary compounds (phenols), polysaccharides, RNA and proteins are eliminated, resulting in purer DNA [18]. Good quality of DNA, can be further used for southern-blotting or for PCR reaction.
-
IV. CONCLUSION
Fresh leaves samples of Plumeria sp. proved to be better source for DNA extraction. DNA solution extracted from fresh material has transparent colour. The DNA concentrations extracted from fresh leaf were between 70300 ng/µl, while from fresh flower were between 0 – 40 ng/µl. DNA extracted from silica dried leaf sample has brown colour and need to purify to eliminate the brown colour.
REFERENCES
-
[1] Van Der Heijden, R., D.I. Jacobs, W. Snoeijer, D. Hallard, R. Verpoorte. 2004. The Catharanthus alkaloids: Pharmacognosy and biotechnology. Curr. Med. Chem.11:607-28
-
[2] Mahadani, P., G.D. Sharma, S.K. Ghosh. 2013. Identification of Ethnomedicinal Plants (Rauvolfioideae: Apocynaceae) through DNA Barcoding from Northeast India. J. Phcog. Mag. 35: 255-263
-
[3] Pipan, B., M.M. Zupančič, E. Blatnik, P. Dolničar, V. Meglič. 2018. Comparison of Six Genomic DNA Extraction Methods for Molecular Downstream Applications of Apple Tree (Malus X
domestica). Cogent Food Agric.
4:1, DOI: 10.1080/23311932.2018.1540094
Journal of Advances in Tropical Biodiversity and Environmental Sciences 3(1): 8-11, February 2019 (p-ISSN: 2549-6980)
DOI: 10.24843/atbes.v03.i01.p03 Available online at: https://ojs.unud.ac.id/index.php/ATBES/article/view/48682
-
[4] Cheng. F.S., S.K. Brown, N.F. Weeden. 1997. A DNA Extraction Protocol from Various Tissues in Woody Species. Hort. Sci. 32(5):921-922.
DOI: 10.21273/HORTSCI.32.5.921
-
[5] Attitalla, I.H. 2011. Modified CTAB Method for High Quality Genomic DNA Extraction from Medicinal Plants. Pakistan J. Biol. Sci, 14: 998-999
-
[6] Abbasi, S., S. Afsharzadeh 2016. An Efficient and Simple CTAB Based Method for Total Genomic DNA Isolation from Low Amounts of Aquatic Plants with a High Level of Secondary Metabolites. Progress Biol. Sci. 6 (1): 95-106
-
[7] Doyle, J..J. and J.L. Doyle. 1990. A Rapid DNA Isolation Prosedur for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 19: 11-15.
-
[8] Sahu, S.K., M. Thangaraj, K. Kathiresan. 2012. DNA Extraction Protocol for Plants with High Levels of Secondary Metabolites and Polysaccharides without Using Liquid Nitrogen and Phenol. ISRN Mol. Biol. 2012. https://doi.org/10.5402/2012/205049.
-
[9] Arruda, S.R, D.G. Pereira, M.M. Silva-Castro, M.G. Brito A.M. Waldschmidt. 2017. An Optimized
Protocol for DNA Extraction in Plants with a High Content of Secondary Metabolites, Based on Leaves of Mimosa tenuiflora (Willd.) Poir. (Leguminosae). Genet. Mol. Res. 16 (3): gmr16039063
-
[10] Henry, R.J. 2001. Plant DNA Extraction, in RJ Henry (ed.), Plant Genotyping: the DNA Finger-
11 printing of Plants, CABI, Oxford UK, pp. 239-249.
-
[11] Maniatis, T., E.F. Fritsch, J. Sambrook.
-
1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory.
-
[12] Farooque, A.M.D., A. Mazunder, S. Shambhawee, R. Mazumder. 2012. Review on Plumeria acuminata. Int. J. Res. Phar. Chem. 2:2
-
[13] Porebski, S., L.G. Bailey, B.R. Baum. 1997. Modification of CTAB DNA Extracions Protocol for Plants Cotaining High Polysacharide Polyphenol Components. Plant Molec Biol. Rep. 15:8-15
-
[14] Konde, S. 2007. Preparation of High-Silica Zeolite Beads from Silica Gel. US: Chemical Engineering, Worcester Polytechnic Institute.
-
[15] Male. A.S., F. Kato, C.M. Mukankusi. 2018. DNA Extraction from Silica Gel Preserved Common Bean (Phaseolus vulgaris L.) Laves. Genet. Mol. Res. 17 (4) DOI: http://dx.doi.org/10.4238/gmr16039926
-
[16] Mathew, K.M., S. Jose, Y.S. Rao, U. Gupta, J. Thomas 2014. Optimization of Genomic DNA Extraction from Fresh and Dry Leaves of Large Cardamom (Amomum subulatum Roxb.) for Diversity Analysis. Indian J. Biotechnol.13(2):221-224
-
[17] Guillemaut, P., L. Maréchal-Drouard 1992. Isolation of Plant DNA: A Fast, Inexpensive and Reliable Method. Plant Mol. Biol. Rep. 10: 60-65.
-
[18] Surzycki, S. 2000. Basic Techniques in Molecular Biology. Springer-Verlag: Berlin, Heidelberg.
Discussion and feedback