CAPSULAR GENOTYPING OF Klebsiella pneumoniae ISOLATED FROM CLINICAL SAMPLES AT SANGLAH GENERAL HOSPITAL IN 2013
on
> JHSM UNUD Journals, Vol. 1 No. 1, February 2017 (DOUBLE-CLICK HERE TO EDIT) <
21
CAPSULAR GENOTYPING OF Klebsiella pneumoniae
ISOLATED FROM CLINICAL SAMPLES AT SANGLAH GENERAL HOSPITAL IN 2013
I Made Sutha Saskara1, Ni Nengah Dwi Fatmawati2, Ni Nyoman Sri Budayanti2, Wahyu Hidayati2 1School of Medicine, Udayana University
Bali, Indonesia suthasaskara@gmail.com
-
2 Department of Clinical Microbiology, School of Medicine, Udayana University, Denpasar, Bali, Indonesia
Klebsiella pneumoniae, one of hospital associated infections agents, become more prevalence in worldwide. It is an opportunistic pathogen that can cause wide spectrum of diseases such as pneumonia and sepsis. Capability of this bacterium in causing diseases is influenced by its virulence factors. Capsule (K) is considered as the major virulence factor responsible in avoiding first defense mechanism of host cells. Almost no study about molecular characterization of capsule is reported in Bali; therefore, this study is aimed to determine the capsular genotype of K. pneumoniae isolated from clinical specimens at Sanglah General Hospital. Samples were collected between January until Desember 2013 in Sanglah General Hospital at Clinical Microbiology Laboratory. Total of 56 samples were examined taken from blood, urine, pus, sputum, and other sources. All K. pneumoniae DNA were then subjected to PCR using specific primer pairs against K1, K2 and K5. The results showed that from 56 K. pneumoniae clinical isolates only 12 (21.4%) were PCR positive, and all (100%) of them were positive for K2 capsule gene. Ten (83.3) of them were from blood, 1 (8.3%) from sputum, and 1 (8.3%) from other specimens. Finding of K2 capsule gene in most K. pneumoniae clinical isolates at Clinical Microbiology Laboratory in Sanglah General Hospital might figure out the relationship of capsular type with severity of diseases. However, further study of capsule in Bali with higher isolates number will help in understanding of pathogenicity of K2 capsule in order to treat the infections itself.
Keywords: Capsule, genotyping, Klebsiella pneumoniae, PCR.
Since its discovery by Carl Friedlander in 1882, K. pneumoniae has become common bacteria infection agents in human.1 In some developing countries, infectious disease trends tend to increase from time to time. From many bacteria classification, negative-gram bacteria has high number of cases and considered as an major infectious agents.
Normally K. pneumoniae is a commensally found in human intestinal system but it can cause considerable morbidity if it is found as agent of disease outside its normal flora such as sepsis, urinary tract infection, or pneumonia. Systemic K. pneumoniae infections could be hard to treat because of its capsule characteristic in preventing human neutrophil phagocytosis process. So far
there are 77 K antigen and 9 O antigen capsular antigens has been identified. K antigen has specific role in its virulence and K1 is the most pathogen among all. 2,3
There is no previous study of K. pneumoniae K1, K2, and K5 capsule gene in Bali. By characterizing the molecular gene of capsules, it may provide us with an insight into better understanding of K. pneumoniae infection pathogenesis. Based on that situation, to serve as a data of K. pneumoniae capsule spread in Bali this study aims to identify the molecular gene characteristic of K. pneumoniae capsule in Bali isolated from Microbiology installation in Sanglah General Hospital in 2013.
-
II. mATERIALS AND METHODS
-
A. Bacterial Strains
Fifty-six K. pneumoniae isolated from clinical specimens examined in Clinical Microbiology Laboratory in Sanglah
General Hospital during January-Desember 2013 were used
in this study. Isolates were taken from blood (23), urine (17), pus (1), sputum (11) and other sources (4). Glycerol stocks of K. pneumoniae isolates were grown onto 5% sheep blood agar plates and incubated at 37oC for 18-24 hours, aerobically.
-
B. Polymerase Chain Reaction
Bacterial colonies were collected for DNA isolation (High Pure PCR Template Preparation Kit, Roche, Germany) and then subjected to PCR for detection of capsular K1, K2, and K5 genotype of K. pneumoniae, which used specific primer as detailed by Turton et al., (2008) in table 3.1. PCR were carried out in 25 μL volumes
(Go Taq Green Master Mix, Promega) that 2 μL extracted
DNA and 0,2 μM of each genotype primer. PCR conditions were: pre-denaturation 94oC for 5 minutes, followed by 30 cycles of 94oC denaturation for 30 seconds, annealing 50oC for 45 seconds, extension 72oC for 90 seconds and a final extension at 72oC for 6 minutes.
The 2% agarose gel in TBE buffer was used for electrophoresis. Gels were run at a constant voltage of 100 V for 1 hour, stained in 2 μg/mL ethidium bromide for 10 minutes and photographed under UV by Gel-Doc (Biorad). The expected PCR products were 1283 bp band for K1, 641 bp band for K2, and 280 bp band for K5.
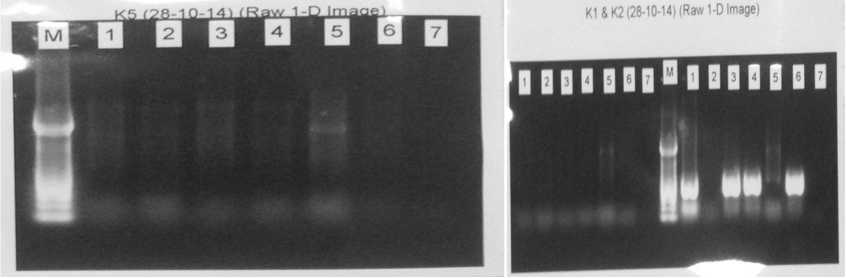
Fig.. 1 Uniplex PCR Running Result. This sample showed 612 bp band formed indicating positive result for K2 in sample number 1,3,4, and 6. Meanwhile there is no band formed in sample of K1 and K5 which indicate negative result respectively.
-
III. RESULTS AND DISCUSSION
The results showed that from 56 K. pneumoniae clinical isolates, only 12 (21.4%) were PCR positive, and all (100%) of them were positive for K2 capsule gene. Ten (83.3) of them were from blood, 1 (8.3%) from sputum, and 1 (8.3%) from other specimens.
TABLE 1
DISTRIBUTION OF CAPSULAR GENOTYPE OF K. PNEUMONIAE BASED ON SPECIMEN TYPES
|
Specimen type (N) |
Capsular type (N) | ||
|
K1 |
K2 |
K3 | |
|
Blood (23) |
0 |
10 |
0 |
|
Urine (17) |
0 |
0 |
0 |
|
Sputum (11) |
0 |
1 |
0 |
|
Pus (1) |
0 |
1 |
0 |
|
Others (4) |
0 |
0 |
0 |
Twelve isolates were found to be K2-positive capsules, in which mostly were taken from bacteremia patient. This result is different compared to Liao et al., (2011) which they conducted a study in 255 bacteremia patients in Taiwan. They found that in Taiwan there is a increasing trend of K1-capsule carrying K. pneumoniae that causes liver abscess. According to their study, 41 (18,2%) positive for K1 genotype, 37 (16,4%) positive for K2, 15 (6,7%) were positive for K57, and 8 (3,6%) positive for K54. It is
not a doubt that K1 and K2 are major capsule genotypes in Taiwan with balanced number on both sides. K1 genotype capsule has lower hospital mortality number compared to K2 (14.6% vs. 43.2%; OR = 0.23, 95% CI 0.08–0.67, p = 0.007), but has a higher chance developing into a liver abscess.4
Meanwhile from isolates that were taken from urine, there was once again a difference in the result compared to a study by Feizabai et al., (2013) in Teheran. They analyzed 89 isolates to detect K1 and K2 genotype capsules. They found 9 (10%) isolates were positive for K1 genotype in urine specimen and 1 (1%) was positive for K1 genotype in a samples taken from trachea. For K2 genotype, they found 11 (12,3%) were positives in urine specimens and 2 (2%) from trachea specimens.5 This is contrary with what we have found in present study, which found no positive isolates for K1, K2, or K5 genotypes.
K2 genotype capsule analyzed prior to its infection history has equal number of case percentage in hospital (9%, 7/75) and community (11%; 16/141). This become a main concern which K2 genotype capsule could be a dangerous threat in community as it is in hospital environment. Compared to K1 genotype capsule, this capsule has tendency to rarely cause a significant number of cases in hospital setting. Such high number of K2-positive genotype capsule may be related to a lot of factors such as genetic predilection (susceptibility of Asians to liver
abscess), underlying diseases (for example, higher prevalence of chronic hepatitis B virus infection in Taiwan), social factors (different foods or cultural practices), and economic factors (for example, access to healthcare, antimicrobial drug usage) may be responsible for the different manifestations of serious K. pneumoniae infection observed in different regions. Despite these other possibilities, our experimental studies suggest that the differences in clinical features arise from differences in the virulence of individual microorganisms.6
Graded to its virulence capability, K2 genotype capsule virulency is almost as virulent as K1 genotype capsule. According to a study conducted by Lin et al., (2014) in 1757 liver abscess patients in Taiwan, K2 genotype capsule is the second highest number cases to be found after K1 genotype capsules. From series of testing the high virulence of K2 genotype capsules is due to its neutrophil and blood serum phagocytosis resistance. But in some cases, where
-
IV. CONCLUSION
Finding of K2 capsule gene in most K. pneumoniae clinical isolates at Clinical Microbiology Laboratory in Sanglah General Hospital might figure out the relationship of capsular type with severity of diseases. However, further study of capsule in Bali with higher isolates number will help in understanding of pathogenicity of K2 capsule in order to treat the infections itself.
Acknowledgment
This work was partly financially supported by Hibah Penelitian Unggulan Udayana Lembaga Penelitian dan Pengabdian kepada Masyarakat (LPPM) Udayana University, Bali, Indonesia under Grant No. 238-8/UN14.2/PNL.01.03.00/2014. Appreciation is dedicated to Ni Wayan Nilawati (Clinical Microbiology Laboratory Sanglah General Hospital staff) for their technical supports.
References
-
[1] Can AD, Alsharif A, Muthiah MP. The Bulging Fissure Sign Of Klebsiella Pneumonia: It's Not A Tumor!. InA48. PULMONARY INFECTIONS: CASE STUDIES (BACTERIAL) 2015 May (pp. A1819-A1819). American Thoracic Society.
K2 genotype capsules didn’t have much adequate resistance to phagocytosis process, it still has high number of virulence. This indicates that maybe another factor contributing to its virulence process but the mechanism it yet to be known.7
From long ago rmpA gene is considered has a significant role in contributing to K. pneumoniae virulence. This gene is often can be found in K1, K2, and non K1 & K2 genotype capsules. This gene has a role in mucoid phenotype regulator. But according to a study by Yeh et al., (2007) the rmpA gene is not a major factor in K. pneumoniae virulence mechanism. In some bacteria, non K1 or K2 genotype capsule which contain rmpA gene did not cause a significant number of virulence compared to K1 or K2 genotype capsule of K. pneumoniae that also contain rmpA gene. This strongly indicate that the genotype of the capsule itself that determine the bacteria virulence.2
-
[2] Yeh, K., Kurup, A., Siu, L., Koh, Y., Fung, C., Lin, J., Chen, T., Chang, F. and Koh, T. (2006). Capsular Serotype K1 or K2, Rather than magA and rmpA, Is a Major Virulence Determinant for Klebsiella pneumoniae Liver Abscess in Singapore and Taiwan. Journal of Clinical Microbiology, 45(2), pp.466-471.
-
[3] Fung, C., Chang, F., Lin, J., Ho, D., Chen, C., Chen, J., Yeh, K., Chen, T., Lin, Y. and Siu, L. (2011). Immune response and pathophysiological features of Klebsiella pneumoniae liver abscesses in an animal model. Lab Invest, 91(7), pp.1029-1039.
-
[4] Liao, C., Huang, Y., Lai, C., Chang, C., Chu, F., Hsu, M., Hsu, H. and Hsueh, P. (2011). Klebsiella pneumoniae Bacteremia and Capsular Serotypes, Taiwan. Emerg. Infect. Dis., 17(6), pp.1113-1115.H. Poor, An Introduction to Signal Detection and Estimation. New York: Springer-Verlag, 1985, ch. 4.
-
[5] Feizabadi, M., Raji, N. and Delfani, S. (2013). Identification of Klebsiella pneumoniae K1 and K2 Capsular Types by PCR and Quellung Test. Jundishapur Journal of Microbiology, 6(9).
-
[6] Yu, V., Hansen, D., Ko, W., Sagnimeni, A., Klugman, K., von Gottberg, A., Goossens, H., Wagener, M. and Benedi, V. (2007). Virulence Characteristics of Klebsiella and Clinical Manifestations of K. pneumoniae Bloodstream Infections. Emerg. Infect. Dis., 13(7), pp.986-993.J. Jones. (1991, May 10). Networks (2nd ed.) [Online]. Available: http://www.atm.com
-
[7] Lin, J., Koh, T., Lee, N., Fung, C., Chang, F., Tsai, Y., Ip, M. and Siu, L. (2014). Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathogens, 6(1), p.21
Discussion and feedback