ISOLATION AND ABILITY TEST OF PHENOL-DEGRADING BACTERIA FROM THE LIQUID WASTE IN BHAYANGKARA HOSPITAL, DENPASAR CITY, BALI, INDONESIA
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 9 No. 1 • September 2021
eISSN: 2655-9994
pISSN: 2303-3371
https://doi.org/10.24843/IJBB.2021.v09.i01.p06
ISOLATION AND ABILITY TEST OF PHENOL-DEGRADING BACTERIA FROM THE LIQUID WASTE IN BHAYANGKARA HOSPITAL, DENPASAR CITY, BALI, INDONESIA
Shara Yulita Harianja, Made Sritamin*, I Gede Putu Wirawan
Faculty of Agriculture, Udayana University
Jl. PB. Sudirman, Denpasar, Bali 80852, Indonesia
*Corresponding author: igpwirawan@unud.ac.id
ABSTRACT
Phenol is an organic compound that is toxic to humans and the environment. This compound was contained in the hospital liquid waste and petrochemical-based industrial sectors. This study aims to obtain hydrocarbonoclastic bacterial isolates that act as biodegradation agent in waste handling. Samples obtained from the biofilter tank of liquid waste treatment Bhayangkara Hospital Denpasar,Bali, Indonesia then carried out isolation and inoculation of bacteria in selective media NA (Nutrient Agar) containing Phenol. The bacteria obtained was performed the viability test on the media with various Phenol concentration levels, obtained three isolates of bacteria capable of growing to the concentration Phenol 500 mg / L, i.e. isolates IB2, IB5, and IB10. Therefore, it was performed the degradation ability and rate test on those three isolates. After 72 hours of incubation periods, the change of Phenol concentrations was analyzed using the folin-ciocalteau method and examined using the spectrophotometer with 660 nm of wavelength. Based on the analysis of final phenol content, the IB2 isolate has the highest degradation capability in the concentration of 600 mg/L (153,4 mg) with a degradation rate of 2,13 mg/h. IB5 isolates have the highest degradation capability in the concentrations of 700 mg/L (109,45 mg) with a degradation rate of 1,52 mg/h. Meanwhile, IB10 isolates show the lowest ability and degradation rate in both concentrations. Through morphological identification and Gram stain, three isolates belong to the Gram-negative rod-shaped bacteria, which has similarities with the Pseudomonas genus.
Keywords: Bacteria, Degradation, Liquid waste, Phenol
INTRODUCTION
Phenols are components of pollutant compounds often found in medical liquid waste such as hospitals and pharmaceuticals, paper industries, and other petrochemical industries. Phenol is a colorless crystalline substance with a distinctive odor, classified into an aromatic
compound. Phenol is used as an antiseptic and disinfectant ingredient (Juwita et al., 2014; Geng et al., 2006). Phenol waste is classified as hazardous waste due to its high toxicity properties (Lethal Dosage LD 50 = 530 mg/kg (orally in rats). Toxicity Level of Phenol depending on the number
of atoms or molecules attached to its benzene chain. Phenols have been declared a priority pollutant by the US Environmental Protection Agency (EPA) and Canada's National Pollutant Release Inventory (NPRI). This compound can be said to be safe for the environment if its concentration ranges from 0.5 – 1.0 mg/L by KEP No.51/MENLH/10/1995 and the phenol threshold in drinking water raw water is 0.002 mg/L as stated by BAPEDAL (Slamet, 2005). The case of groundwater pollution by phenol compounds results in a decrease in water quality and affects the taste and aroma of the water. Phenols also harm human health depending on exposure and concentration since phenols are corrosive and carcinogens in the human body. This compound can also affect the central nervous system and immune disorders.
The method to minimize phenol pollution in a liquid waste can be made by utilizing microbes such as bacteria as biodegradation agents. Biodegradation occurs because bacteria use contamination compounds as a food source (substrate) and convert them into carbon dioxide or other harmless substances through an enzymatic reaction. From several previous research, there are several bacterial genera involved in the degradation of phenol compounds in
waste and classified into Hydrocarbonoclastic bacteria, including members of the genus Achromobacter, Alcaligenes, Acinetobacter, Arthrobacter, Azotobacter, Flavobacterium, Nocardia, Bacillus cereus, Pseudomonas putida, P. aeruginosa, and Brevibacterium fuscum (Bitton, 2005; Essenberg et al., 2008). Pseudomonas is the most widely reported genus of bacteria that can degrade phenols (Annadurai et al., 2007; Agarry et al., 2008a; Agarry et al., 2008b; Agarry et al., 2008c; Lin et al., 2008; Agarry et al., 2009).
Based on the impact of phenols on liquid waste, this research needs to be done to determine how the influence of bacterial activity in lowering the concentration of phenols in liquid waste so that obtained efforts to treat phenol waste microbiology are safe for the environment and living things. The hypothesis of this study is the securing of bacterial isolates belonging to hydrocarbonoclastic bacteria, namely bacteria that can degrade phenol compounds in isolation from hospital wastewater.
MATERIALS AND METHODS Preparation of Culture Media for Bacteria
There were three types of media used in this research, i.e., LB (Lactose Broth), NB (Nutrient Broth), and NA (Nutrient Agar) instant media with doses of 2.5 g/100 mL, 0.8 g/100 mL, and 2 g/100 mL, respectively. The mixture of each medium was homogenized on a magnetic stirrer for 5 minutes, then heated in the microwave for 15 minutes, and sterilized in an autoclave at a temperature of 121°C for 15 minutes.
Isolation, Characterization, and
Purification of Phenol Degrading Bacteria
Bacteria are isolated from wastewater in the biofilter tank of The Wastewater Treatment Plant (IPAL) of Bhayangkara Hospital Denpasar. Bacterial isolation is carried out using two ways: the incubation treatment of wastewater samples into NB media containing Phenol 50 mg /L in shakers for 48 hours at room temperature at a speed of 150 rpm. The second method is carried out using a multilevel dilution of 10-1 to 10-4. The suspension of both ways, as much as 1 ml, is inoculated by pour plate method on LB media containing 50 mg/L phenol and incubated for 24-48 hours at 28°C until
eISSN: 2655-9994 pISSN: 2303-3371 https://doi.org/10.24843/IJBB.2021.v09.i01.p06
there is a growth of bacterial colonies. Mixed bacterial colonies are grouped and characterized by the morphology of the territory, namely colony shape, colony color, colony edge, and elevation, and further purified in NA media containing 50 mg /L phenol using the streak plate method.
Viability Test of Bacterial Isolate Selected Against Phenols
Viability tests were conducted using five concentrations of multilevel Phenols (100 mg/L, 200 mg/L, 300, 400, and 500 mg/L) in the media of a bacteria incubated at 28 °C for 48 hours and observed colony growth every 24 hours.
Inoculation is carried out using puncture culture as many as nine punctures in the NA media. Then observations are made on the growth of colonies on each punch.
Degradation Capability Test Phenol Isolates Selected Bacteria
The bacterial isolate capability test was selected by cultured 10 ml suspension of each bacterial isolate into 100 ml of NB media on the Erlenmeyer containing test concentrations (600 mg/L and 700 mg/L) in shakers at a speed of 150 rpm for 72 hours at room temperature. There are controls on each test concentration that is a medium that does not have a bacterial
suspension in it. The incubation result is centrifuge for 5 minutes at a speed of 12,000 rpm with a temperature of 4 °C to separate bacteria from phenol solution. The final concentration of Phenol on the supernatant is examined on the UV-VIS
spectrophotometer at a wavelength of 660 nm.
The presentation of degraded phenol levels is calculated using the following equations:
Initial concentration — End concentration
Initial concentration
× 100%
The rate of degradation calculated using the formula:
So - Si
t
Gram Staining of Selected Bacteria
Gram staining of bacteria using several reagents consisting of the paramount dye violet crystals, mordan solution is lugol as a binder, alcohol 98% as a fastener, and safranin a covering dye. Preparations are observed under an electron microscope using a magnification of 1000x. The surface of the bacterial mold is tested using emersi oil to make the appearance of bacterial cells more clearly visible.
Description:
-
S0: Initial phenol content
-
S1: Final phenol content t : Incubation time
by 63 colonies (Figure 1). (A) is a colony of bacteria obtained from samples by incubation method, (B) and (C) is a colony of bacteria obtained from samples by dilution method 10-4.
The characterization of colony morphology is done as an early indication of bacterial purity observed based on the morphology of the colony, obtaining ten isolates that have differences (Table 1).
RESULTS AND DISCUSSION
Bacterial Isolation of Phenol Compound
Degradation
The isolation of phenol-degrading bacteria from biofilter tanks was obtained
Table. 1. The morphological character of colony ten isolates of isolated bacteria
Isolated Colony Morphological
|
Code |
Shape |
Margin |
Elevation |
Color |
|
IB1 |
Irregular |
Lobate |
Raised |
White |
|
IB2 |
Irregular spread |
Lobate |
Flat |
White |
|
IB3 |
Circular |
Entire |
Convex |
White |
|
IB4 |
Circular |
Entire |
Flat |
White |
|
IB5 |
Circular |
Curled |
Flat |
White-clear |
|
IB6 |
Circular |
Undulate |
Convex |
Yellow |
|
IB7 |
Irregular spread |
Lobate |
Raised |
Transparant |
|
IB8 |
Filamentouse |
Curled |
Raised |
Creamy-white |
|
IB9 |
Circular |
Entire |
Concave |
White |
|
IB10 |
Circular |
Entire |
Raised |
White |
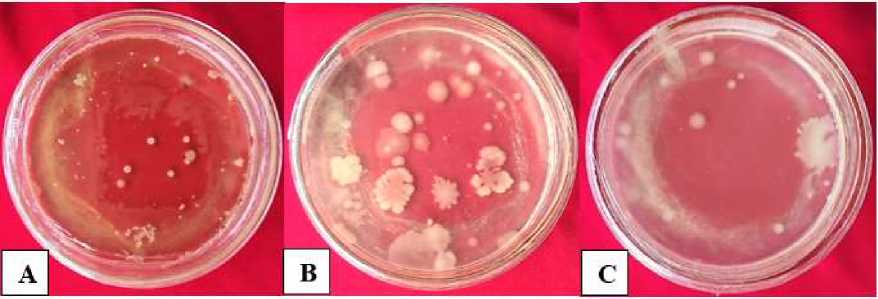
Fig. 1. The whole colony of mixed bacteria isolated from the liquid waste biofilter tank of Bhayangkara Hospital Denpasar
Isolates Selected Bacteria Degrading Phenol Compounds
A viability test of 10 isolated isolates (Table 4.2) showed that each bacterial isolate showed a different growth response in media containing test concentrations. Such growth responses such as the size of
the diameter of the bacterial colony on each puncture, discoloration of the colony, the development of the colony at each incubation period. The difference in colony size of each bacterial isolate in the test media can be seen in Figure 2.
Table. 2. Viability test of 10 isolates of isolated bacteria against phenols
Isolate Phenol Solution Concentration (mg/L)
|
Code |
100 |
200 |
300 |
400 |
500 |
|
IB1 |
• + |
+ |
+ |
° | |
|
IB2 |
• + |
• + |
• + |
• + |
• + |
|
IB3 |
+ |
• + |
° |
- |
- |
|
IB4 |
• + |
• + |
• + |
° |
° |
|
IB5 |
• + |
• + |
• + |
• + |
• + |
|
IB6 |
+ |
- |
- |
- |
- |
|
IB7 |
• + |
• + |
+ |
- |
° |
|
IB8 |
+ |
- |
- |
- |
° |
|
IB9 |
+ |
- |
- |
- |
- |
|
IB10 |
• + |
• + |
• + |
• + |
• + |
Descrption : ° : Isolates of bacteria undergo small colony growth
+ : There are the growth and development of colonies - : No colony growth occurs at such concentrations
• : Bacterial isolates have a large colony size
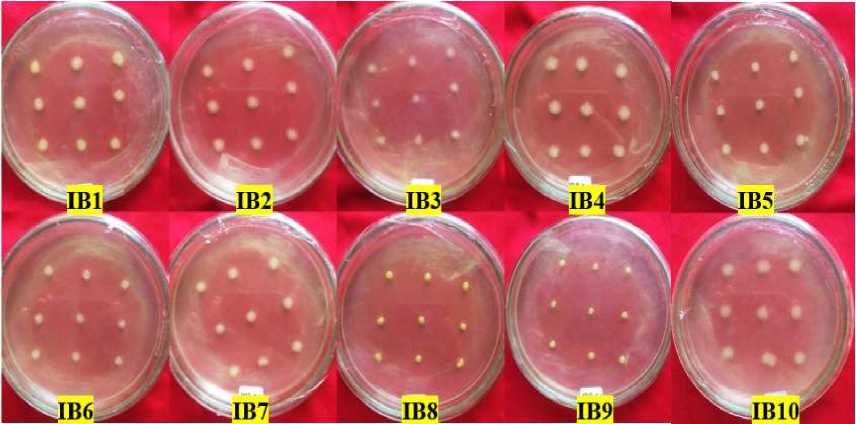
Fig. 2. Bacterial colony growth in 100 mg/L phenol concentration viability
test at 48-hour incubation period
Viability tests of 10 isolated bacterial isolates at initial concentrations (100 mg/L) showed all bacterial isolates could grow well, judging by the uniform colony size of each puncture. But in bacteria with colic codes, IB1, IB2, IB4, IB5, IB7, and IB10 have a larger colony size. This can be
due to isolates of these bacteria better in utilizing phenol compounds as a source of carbon than isolates whose colony growth is more negligible (IB6, IB8, IB9).
The viability test aims to determine the maximum viability of isolates selected in media containing phenols to obtain
bacterial isolates that can grow well because they can utilize phenol compounds for top growth to the highest concentration.
Viability test at a concentration of 300 mg/L obtained as many as seven isolates of bacteria that can grow, but three of them have a small colony diameter growth. At concentrations of 400 mg/L and 500 mg/L, four isolates (IB2, IB4, IB5, and IB10) can continue to grow with large colonies except for IB4 isolates whose territories are shrinking as phenol concentrations in the test medium increase.
Figure 3 shows IB2, IB5, and IB10 isolates capable of growing at all concentration levels. These three isolates have consistent growth starting from initial selection with concentrations of 100 mg/L to 500 mg/L. This is also seen from the increase and size of the colony's enlarged diameter in all puncture holes in the medium. This indicates that the bacteria isolate is tolerant of high phenol concentrations and can make maximum use of phenol as a substrate for its growth.
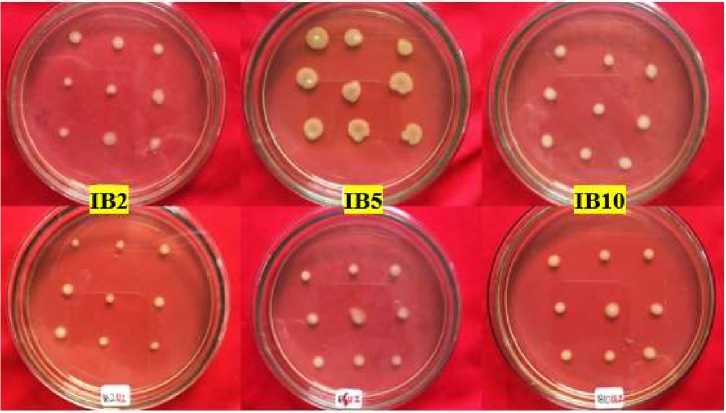
Fig. 3. Bacterial colony growth in viability tests concentrations of 400 mg/L (above) and 500 mg/L (bottom) phenols during the 24-hour incubation period
Test the Degradation Capability of Selected Bacterial Isolates
Phenol compound degradation capability test was conducted against three isolates (IB2, IB5, and IB10) selected in the bacterial isolate viability test against
phenol-containing media with two test concentrations of 600 mg/L and 700 mg/L.
Table. 3. Degraded phenol levels and bacterial isolate degradation rate selected at both test concentrations
|
Isolate Code |
Phenol Degraded at Test Concentration |
Rate of Degradation | ||||
|
600 mg/L |
700 mg/L | |||||
|
mg/L |
(%) |
mg/L |
(%) |
600 mg/L |
700 mg/L | |
|
IB2 |
153,4 |
25,5 |
102,6 |
14,65 |
2,13 |
1,43 |
|
IB5 |
151,5 |
25,25 |
109,45 |
15,63 |
2,10 |
1,52 |
|
IB10 |
116,7 |
19,45 |
63,85 |
9,12 |
1,62 |
0,88 |
|
Control |
106,8 |
17,8 |
43,05 |
6,15 |
1,48 |
0,60 |
Description: Degraded phenols result from a reduction of the initial concentration of phenols with the final concentration of phenols in the test medium after a 72-hour incubation period examined using a spectrophotometer. Control is a test media that does not contain bacterial isolates. (%) is the presentation of phenol degraded from the total initial concentration.
The results of the final phenol level examination on the test media conducted on the spectrophotometer can be seen in Table 3, which shows that the three isolates of selected bacteria can lower the concentration of phenols during the incubation period 72 hours with different abilities. The lower the phenol levels in the test media, the higher the ability of bacteria to degrade, and the rate of degradation increases because the required incubation time is less.
Based on the results of the degradation of selected bacteria contained in the Table. 3, at the test stage, of IB2 bacterial isolate ability, has the best degradation capability and the highest degradation rate of the three isolates, IB2 isolates can lower phenol levels by 153.5 mg/L with a degradation rate of 2.13 mg/h during the 72-hour incubation period. IB10 bacterial isolate is a bacterial isolate that
can lower phenol levels in the lowest I ability test media which is only 116.7 mg L with a degradation rate of 1.48 mg/h.
In the phase II concentration of phenol concentration of 700 mg/L, shown in Table .3, the IB5 bacterial isolate has the highest degradation capability and the most significant rate of degradation of the three isolates tested. IB5 isolates were able to lower phenol levels by 109.45 mg/L with a degradation rate of 1.52 mg/h, in contrast to the test of the ability of the best isolate I in degrading phenol compounds is IB2 isolates, so this proves that IB2 ability to degrade phenol compounds is decreasing at increased phenol concentrations during the same incubation period. The rate of degradation of all three bacterial isolates decreases at higher phenol concentrations.
As the phenol concentration increases, it increases the time it takes to degrade it, thus lowering its degradation
rate. Control treatment or media that does not contain bacteria also decreased phenol levels and not much different from one of the bacterial isolates in the test media. Control in the stage I capability test phenol concentration of 600 mg/L decreased phenol levels by 106.8 mg/L with a degradation rate of 1.48 mg/h during the 72-hour incubation period. In the test of the ability of stage II phenol concentration 700 mg/L, there was a decrease in 43.5 mg/L. This decrease occurred during the shaker during the incubation period that caused the aeration process, which is a manual way of lowering phenol levels (Juwita et al., 2014).
Gram Staining of Bacterial Cells
Gram staining results in this study seen in Figure 4 conducted against three isolates of selected bacteria, namely IB2, IB5, and IB10 bacterial isolates. The three bacteria produce a red color on gramtightening so that it belongs to gramnegative bacteria. Cells do not form spores and stem-shaped bacterial cells or basil suspected to have similarities to the genus Pseudomonas.
Gram-negative bacteria are known to have higher resistance or resistance to antibiotic activity, so in locations exposed to compounds that have an antibiotic action, such as phenols, are often found to be Gram-negative bacteria.
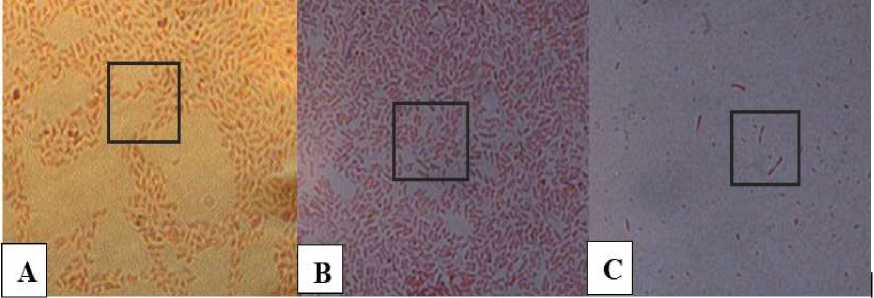
Fig. 4. Gram Staining of selected bacterial isolates (A) IB2; (B) IB5; (C) IB10; observed on a microscope with a magnification of 1000x. (the shape of bacterial cells is visible on the box).
CONCLUSION
Isolation obtained 63 colonies of bacteria that can grow on selective phenol media and grouped into 10 isolates based
on their morphological character. Three isolates (IB2, IB5, IB10) can grow up to a concentration of 700 mg/L of phenols that are Gram-negative bacteria with no spores
and are rod-shaped, which are thought to have similarities to the genus Pseudomonas.
IB2 isolates have the highest degradation capability at phenol concentrations of 600 mg/L, IB5 isolates have the highest degradation capability at phenol concentrations of 700 mg/L. In contrast, IB10 isolates have the lowest capabilities at both test concentrations.
REFERENCES
Agarry, S. E., Audu, T. O. K., & Solomon,
B. O. (2009). Substrate inhibition kinetics of phenol degradation by Pseudomonas fluorescence from steady state and wash-out data. International Journal of Environmental Science &
Technology, 6(3), 443-450.
Akmal. (2010). Biodegradasi Fenol Limbah Cair Industri Tekstil oleh Candida tropicalis. Jurnal skripsi. Departemen Biolokimia. Fakultas Matematika dan Ilmu Pengetahuan Alam. Institur Pertanian Bogor.
AMINI, B. N., Hassanshahian, M., &
Khoshrou, S. M. R. (2014). Isolation and characterization of phenol degrading bacteria from Persian Gulf.
Badan Pusat Statistika. (2018). Produksi Fenol. Publikasi BPS. Jakarta.
Bergey, D. H., & D.R, Boone. (2009). Bergey’s Manual of Systematic Bacteriology. Vol.3, Ed.2. Springer Science-Business Media, New York.
Bitton, G. (2005). Wastewater microbiology. John Wiley & Sons.
Fitriana, F., Baharuddin, M., & Sappewali, S. (2016). Isolasi dan Identifikasi Bakteri Pendegradasi Fenol yang
Bersumber Dari Danau Tempe Kabupaten Wajo Sulawesi
Selatan. Al-Kimia, 4(2), 132-141.
Juwita, R., Fibriarti, B. L., & Roza, R. M. (2014). Isolasi dan Karakterisasi Bakteri Pendegradasi Fenol dari Limbah Cair Rumah Sakit Umum Daerah (RSUD) Arifin Achmad Pekanbaru (Doctoral dissertation, Riau University).
Lestari, D. M., Mahmudati, N., Sukarsono, S., Nurwidodo, N., & Husamah, H. (2018). Aktivitas Antioksidan Ekstrak Fenol Daun Gayam (Inocarpus fagiferus
Fosb). Biosfera, 35(1), 37-43.
Mohite, B. V., Jalgaonwala, R. E., Pawar, S., & Morankar, A. (2010). Isolation and characterization of phenol degrading bacteria from oil contaminated soil. Innovative Romanian Food Biotechnology, 7, 61.
Nweke, C. O., Okpokwasili, G. C., & Harcourt, P. (2014). Kinetics of growth and phenol degradation by Pseudomonas species isolated from petroleum refinery wastewater. Int. J. Biosci, 4, 28-37.
Rocha, L. L., de Aguiar Cordeiro, R., Cavalcante, R. M., do Nascimento, R. F., Martins, S. C. S., Santaella, S. T., & Melo, V. M. M. (2007).
Isolation and characterization of phenol-degrading yeasts from an oil refinery wastewater in
Brazil. Mycopathologia, 164(4),
183-188.
Sachan, P., Madan, S., & Hussain, A. (2019). Isolation and screening of phenol-degrading bacteria from pulp and paper mill effluent. Applied Water Science, 9(4), 1-6.
Yulvizar, C. (2011). Efektivitas Pengolahan Limbah Cair Dalam Menurunkan Kadar Fenol Di Rumah Sakit Umum Daerah Dr. Zainoel
Abidin (Rsudza) Banda Aceh. Jurnal
Biologi Edukasi, 3(2), 9-15.
59
Discussion and feedback