PRIMER DESIGN OF CVPDr DNA FRAGMENT SEQUENCES THAT AMPLIFY SPECIFIC FRAGMENTS TO DISTINCT THE RESISTANT FRAGMENT FROM Triphasia trifolia (Burm. F.) P. Wils. AND THE SUSCEPTIBLE FRAGMENT FROM Citrus nobilis Lour.
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 7 No. 2 • April 2020 eISSN: 2655-9994 pISSN: 2303-3371 https://doi.org/10.24843/IJBB.2020.v07.i02.p05
PRIMER DESIGN OF CVPDr DNA FRAGMENT SEQUENCES THAT AMPLIFY SPECIFIC FRAGMENTS TO DISTINCT THE RESISTANT FRAGMENT FROM
Triphasia trifolia (Burm. F.) P. Wils. AND THE SUSCEPTIBLE FRAGMENT FROM Citrus nobilis Lour.
Ni Made Ayuratih Utami*, I Gede Putu Wirawan, and I Ketut Suada Department of Agroecotechnology, Faculty of Agriculture, Udayana University *Corresponding author: ayuratihutami@gmail.com
ABSTRACT
CVPDr is a DNA fragment that indicates that plants are resistant to CVPD. Previous research using primers that amplified 841 bp CVPDr fragment was able to amplify the fragment from Triphasia trifolia that considers being a resistant plant, Citrus aurantifolia var. seedless which considers being a tolerant plant, and some susceptible citrus plants to CVPD disease. In this study, we designed some primers that amplified only CVPDr DNA fragment from T. trifolia which consider as the resistant plant and a primer that amplified only DNA fragmen from Citrus nobilis which consider as the susceptible citrus plants. The primers for CVPDr on T. trifolia are TCATCTGCATGGGATACC for forward primer and
GCCTTGAGCTTGTAAGTG for reverse primer which turned out to amplify the DNA of T. trifolia and also the C. nobilis cultivar Denpasar and only succeeded in not amplifying the C. nobilis cultivar Gianyar. The primers for CVPDr on C. nobilis are
GAATGGCTTAGCAGAAAGG for forward primer and GGTTGTAGATGGACATAGG for reverse primer turned out can not only amplify the DNA C. nobilis but also amplify T. trifolia.
Keywords: CVPDr fragment, Primer Design, Citrus nobilis, Triphasia trifolia
INTRODUCTION
Citrus fruit production in Bali has fluctuated over the past five years. In 2016, citrus production in Bali from the previous year decreased by 45,133 tons based on various factors decreasing citrus production (Badan Pusat Staristik, 2018), one of which was an attack of Citrus Vein Phloem Degeneration (CVPD). Some popular plants, some non-commercialized citrus plants and some citrus relatives, are proven to be resistant to CVPD.
CVPD is caused by Liberibacter asiaticus which develops in shoots and is transmitted by vector insect Diaphorina citri (Tirtawijaya, 1983; Jaqoueix et al., 1994; 1996; Wirawan, et al., 2004). In addition to oranges, CVPD can be transmitted to several members of the oranges (Rutaceae) tribe such as Poncirus trifoliata (L) Raf, Kemuning / Murraya paniculata (L) Jack, Swinglea glutinosa and Clausena indica.
Martasari et al. (2004), stated that many citrus farmers complained about the condition of their oranges which were
attacked by various diseases both by bacteria and viruses. All citrus plants cultivated are susceptible to CVPD disease attacks (Mahayani, 2013). In several cases several types of citrus plants were reported, notably commercialized citrus plants and some of their relative plants, known to be resistant to CVPD. Among them are seedless lime, Tahiti lime, Triphachia trifolia (kinkit orange), and Poncirus trifoliata (karatachi). Citrus plants that are resistant to CVPD are thought to contain genes that produce a trait that is able to break the pathogenic CVPD infection or is able to resist transmission of pathogens carried by vector insects (Wirawan et al., 2004).
Previous studies was done using DNA fragments isolated using RAPD and mutations from T. trifolia, these fragments were indicated to be resistant factors to CVPD disease and then referred to as CVPDr DNA fragments. Yuniti et al. (2018) found polymorphisms in CVPDr DNA fragments in several citrus species (Citrus, spp) and 12 types of citrus plants that were sampled were plants that had CVPDr DNA fragments. Mutations that occur in some CVPDr DNA fragments cause changes in bases in DNA fragment sequences so that repairing sequences of some of these plants can be distinguished according to their level of sensitivity to CVPD disease.
The purpose of this study is to develop primers that only amplify DNA fragments from resistant plants (T. trifolia) and susceptible plants (Citrus nobilis). This primer is expected to be a specific primer that can be used to detect CVPDr in resistant plants (T. trifolia) and susceptible plants (Citrus nobilis).
MATERIALS AND METHODS
The study was conducted in two stages, namely in silico and in vitro. Research conducted in silico or using a computer is a bionformatics research to design the desired primer.
The results of C. nobilis DNA sequencing from seven regions in Bali and T. trifolia DNA (Yuniti et al., 2018) were converted to FASTA format. The sequences in the FASTA format are then aligned using the Ugene (Okonechnikov, et al., 2012) application with the MUSCLE algorithm. Sequences are sought for conservative regions which are then used as templates for primer designs.
The primer design is done with in silico method using the Clone Manager program through several stages. First install the Clone Manager Pro 9 Program. Prepare the sequential data in FASTA format. Open the Clone Manager Pro 9 program, select the primer menu on the menu bar, then choose design. In the dialog box, enter the desired
primer and reverse primer lengths (18-24 bases); Click the criteria menu then enter the desired primer parameters. Then select next. Enter the data sequence that has been previously saved and the target region is equipped with the desired base position as the start and end of the target region, select finish. In the dialog box primer results will appear based on the primer criteria and sorted by rank.
The Clone Manager program will display several primer candidates according to the required criteria. The best primers candidates then tested for their annealing place using the Ugene application.
In vitro research was carried out after the primer design was completed, which is
the primer test using the PCR method in the laboratory.
RESULTS AND DISSCUSSION
Triphasia trifolia Primer
The CVPDr fragment sequence from kinkit orange is input into the Clone Manager application along with the desired primer parameters. The Clone Manager application then display several suitable primer candidates and sorted by rank. There are five candidate pairs of T. trifolia primers who fit the proposed criteria. Information about the five primer pairs can be seen in table. 1.
Table. 1 T. trifolia primers candidate from the Clone Manager Program
|
Primer |
Jenis |
Primer sequence |
GC |
Tm |
Length |
3’ End |
Any Dimer |
Product |
|
Primer |
% |
(oC) |
Dimer |
Length | ||||
|
Primer |
F |
TCATCTGCAT- |
50 |
57 |
18 |
2 |
4 |
302 |
|
Kinkit 1 |
R |
GGGATACC GCCTTGAGCT- TGTAAGTG |
50 |
57 |
18 |
1 |
4 | |
|
Primer |
F |
GTCATCTGCA- |
52 |
59 |
19 |
2 |
4 |
303 |
|
Kinkit 2 |
R |
TGGGATACC GCCTTGAGCT-TGTAAGTG |
50 |
57 |
18 |
1 |
4 | |
|
Primer |
F |
GGATTGGTCA- |
50 |
60 |
20 |
2 |
3 |
401 |
|
Kinkit 3 |
R |
GCCTACAAAC GCCTTGAGCT-TGTAAGTG |
50 |
57 |
18 |
1 |
4 | |
|
Primer |
F |
GGTCAGCCTA- |
50 |
60 |
20 |
2 |
4 |
396 |
|
Kinkit 4 |
R |
CAAACTTTGG GCCTTGAGCTT-GTAAGTG |
50 |
57 |
18 |
1 |
4 | |
|
Primer |
F |
GGTCATCTGCA- |
55 |
62 |
20 |
2 |
4 |
304 |
|
Kinkit 5 |
R |
TGGGATACC GCCTTGAGCTT-GTAAGTG |
50 |
57 |
18 |
1 |
4 |
From the five pairs of primer candidates produced by Clone manager suite 9, primers were selected that best suit their needs. The selected forward kinkit primer then written as F-Kinkit and the reverse primer is written as R-Kinkit.
F-Kinkit: TCATCTGCATGGGATACC R-Kinkit: GCCTTGAGCTTGTAAGTG
The results of the analysis and primer information can be seen in Fig. 1 and Fig. 2. Based on these pictures it is known that the primer pair has fulfilled good primer criteria.
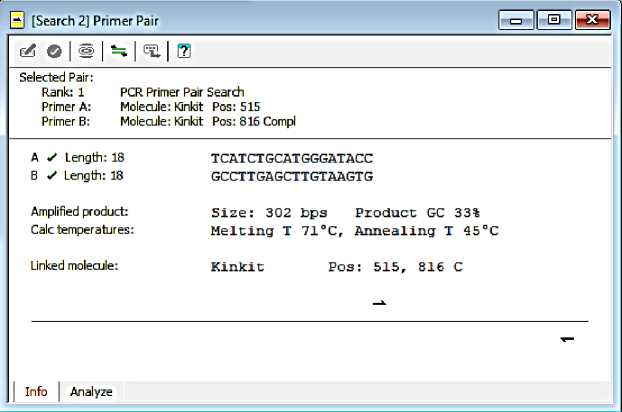
Fig. 1. F-Kinkit and R-Kinkit primer information
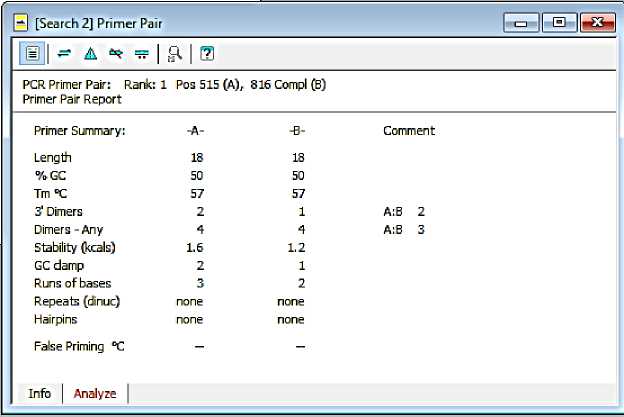
Fig. 2. The results of the analysis of the primer design of F-Kinkit and R-Kinkit
The length of the F-Kinkit and R-Kinkit is 18 bases. Each has a GC percentage of 50% and Tm 57̊C. The general rule that is followed by most primer design programs is to use a base percentage of G and C between 40% to 60% (Dinda Eling, 2014).
Primers may not contain complementary base sequences (palindromes) on the same strand. This can result in the formation of hairpins. The primers will naturally fold and produce unproductive priming. The best primers chosen are primers that do not or have little hairpin structure, dimers, and cross dimers (Breslauer et al., 1986).
Based on the analysis results in the program (Figure 4.2), the two primers do not form a hairpins structure. The selected primer pair has a dimer at the end of 3 'ie two on the primer forward and one on the reverse
primer. The other dimers formed each bind four base strands. Cross dimers are also formed between the two primers, which are at the end of 3 'by two bases and the other dimer by five bases.
Primer attachment position was again tested using the Eugene program. Primers that have been made stick to the sequences of Kinkit Oranges (T. trifolia) and Denpasar Siem orange (C. nobilis). The position of the primer attachment in each sequence is as follows:
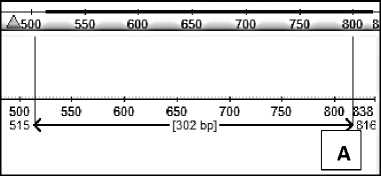
Fig. 3. Position of primer attachment to (A) Kinkit Oranges (T. trifolia) and (B) Denpasar Siem Oranges (C. nobilis)
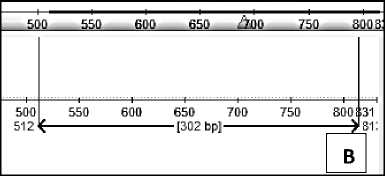
The primers attach to the Triphasia trifolia sequences and Citrus nobilis cul. Denpasar because the two oranges have a similar sequence of bases.
C. nobilis Primer
Primer for C. nobilis was obtained after aligning seven samples of C. nobilis
cultivar in several regions in Bali so that the same base sequence can be seen. The sequence is then used as a template for primer design. Data sequence of the seven samples of siem orange has different lengths, so the extra bases on the front and back of the sequence are removed so that the sequences of the seven samples are the same length. There were 733 bases after the
sequence length of the seven sampled samples were then analyzed with CM suite 9 to obtain the desired primer pair.
There are 59 primer pairs that match the proposed criteria. Four of the 59 primer pairs were ranked first based on ratings in the Clone Manager application. Primer information with the first rank can be seen in table. 2.
Table. 2 C. nobilis primers candidate from the Clone Manager Program
|
Primer |
Jenis Primer |
Primer sequence |
GC % |
Tm (oC) |
Length |
3’ End Dimer |
Any Dimer |
Product Length |
|
Primer Siem 1 |
F |
AGAATGGCTTAG-CAGAAAGG |
45 |
59 |
20 |
1 |
3 |
144 |
|
R |
TGGTTGTAGATG-GACATAGG |
45 |
58 |
20 |
1 |
3 | ||
|
Primer Siem 2 |
F |
GAATGGCTTAGC-AGAAAGG |
47 |
57 |
19 |
1 |
3 |
142 |
|
R |
GGTTGTAGATGG-ACATAGG |
47 |
56 |
19 |
1 |
3 | ||
|
Primer Siem 3 |
F |
GAATGGCTTAGC-AGAAAGG |
47 |
57 |
19 |
1 |
3 |
189 |
|
R |
GTTTGTAGGCTG-ACCAATC |
47 |
57 |
19 |
2 |
3 | ||
|
Primer Siem 4 |
F |
AGAATGGCTTAG-CAGAAAGG |
45 |
59 |
20 |
1 |
3 |
191 |
|
R |
AGTTTGTAGGCT-GACCAATC |
45 |
59 |
20 |
2 |
3 |
F-Siem: GAATGGCTTAGCAGAAAGG R-Siem: GGTTGTAGATGGACATAGG
Compared to the first and fourth primer pairs, the selected primer pairs have a higher GC percentage of 47%. Tm F-Siem is 57̊C, while R-Siem is 56̊C. The primer pair has a Tm difference of 1̊C which is tolerable.
The primer pair does not form a hairpin structure. Each primer has a dimer at the end of 3 'of one base and the other dimer of three bases. Cross dimers are also expected to be formed, which is one end 3 'and three bases complementary to each other.
|
0 [Search 5] Primer Pair ∣∙o-∣∣⅛∣∣∣⅛∣ | ||
|
^ © I S 11⅞ I ¾ I 0 | ||
|
Selected Pair: Rank: 1= PCR Primer Pair Search Primer A: Molecule: daerah Pos: 193 PrimerB: Molecule: daerah Pos: 334Compi | ||
|
A Z Length: 19 GAATGGCTTAGCAGAAAGG B Z Length: 19 GGTTGTAGATGGACATAGG Amplifiedproduct: Size: 142 ⅛≡ Product GC 35% Calctemperatures: Melting T 69°C, Annealing T 43°C Linkedmolecule: daeιaħ Po≡: 193, 334 C Infb I Analyze | ||
Fig. 4. Primer information of F-Siem dan R-Siem
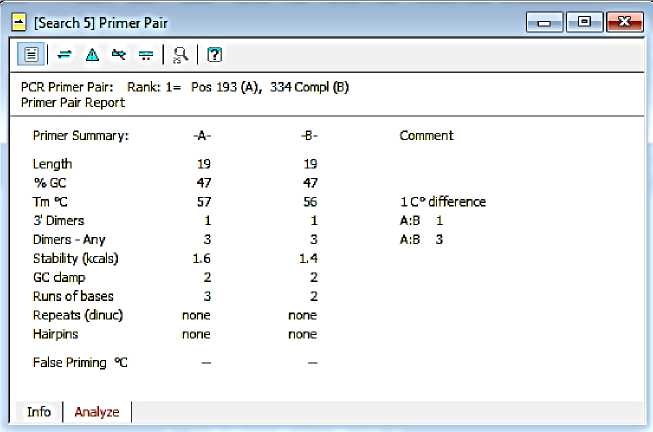
Fig. 5. The results of the analysis of the primer design of F-Siem and R-Siem
The primer attachment sites for each C. nobilis sample in the Ugene application are as follows:
Table. 3 Primer attachment sites F-Siem and R-Siem on each sample C. nobilis
Orange cultivar Attachment sites Product length
|
Siem |
193-334 |
142 |
|
Siem Denpasar |
244-385 |
142 |
|
Siem Buleleng |
235-376 |
142 |
|
Siem Gianyar |
240-381 |
142 |
|
Siem Payangan |
202-343 |
142 |
|
Siem Pecatu |
236-377 |
142 |
|
Siem Tabanan |
249-390 |
142 |
Primer Design Test Results by PCR (Polymerase Chain Reaction) Method
Pair of kinkit primer and siem orange primer design results were tested to find out whether the primer succeeded in amplifying the DNA of oranges that had been isolated. Testing is done by PCR method. Each primer was tested on the three DNA samples namely DNA Triphacia trifolia, Citrus nobilis cul. Denpasar, and Citrus nobilis cul. Gianyar.
DNA isolation from the leaves of siem orange and kinkit orange was carried out to test the primer that had been made.
Total DNA isolation was carried out using the Mini Genomic DNA Kit from Genaid. The success of the DNA isolation stage can be seen from the visualization of agarose gel electrophoresis on the UV transilluminator (Fig. 6). The success of the total DNA isolation process is evidenced by the presence or absence of DNA bands in agarose gels during electrophoresis.
Fig. 6. Visualization of the total DNA isolation results of T. trifolia, C. nobilis cul. Gianyar, and C. nobilis cul. Denpasar
Total DNA
The PCR results were then electrophoresed on 1% agarose gel using a 1% TAE buffer solution. Warming is done for 20 minutes (100 volts). The agarose gel is then transferred to a UV transilluminator for
PCR DNA bands. The presence or absence of DNA bands indicates the primer success in amplifying the target fragment needed. Visualization of PCR results using F-Kinkit and R-Kinkit primers can be seen in Fig. 7.

Fig. 7. The results of DNA amplification using F-Kinkit and R-Kinkit primers (1) C. nobilis cul. Gianyar, (2) C. nobilis cul. Denpasar, and (3) T. trifolia.
The primer pair F-Kinkit and R-Kinkit succeeded in amplifying the DNA of C. nobilis cul. Denpasar, and T. trifolia. DNA bands are seen in lines two and three which prove that DNA was successfully amplified by the primer pairs that had been
designed. Lane one filled with C. nobilis DNA DNA solution. Gianyar did not show the DNA band (Fig. 7). DNA is not amplified according to in silico tests using the Ugene application.

Fig. 8. The results of DNA amplification using F-Siem and R-Siem primers (1) C. nobilis cul.
Gianyar, (2) C. nobilis cul. Denpasar, and (3) T. trifolia.
Primer F-Siem and R-Siem amplified all three DNA samples. The primer is expected not to apply T. trifolia DNA. The
amplification of T. trifolia DNA is probably due to the high level of homology of T. trifolia and C. nobilis DNA. The high level
of homology between genes allows the primer to amplify several genes, although there are differences in several nucleotides (Kamel, 2003).
CONCLUSION
In this study it can be concluded that Primer design results for amplifying CVPDr DNA fragments in the plant of kinkit (T. trifolia) namely TCATCTGCATGGGATACC primer primer and reverse primer
GCCTTGAGCTTGTAAGTG turned out to amplify the DNA of kinkit orange and also the siem orange cultivar Denpasar. The primer only succeeded in not amplifying the Gianyar cultivar siem orange. Primer design results to amplify CVPDr DNA fragments on the plant of siam (C. nobilis), namely GAATGGCTTAGCAGAAAGG forward primer and reverse primer
GGTTGTAGATGGACATAGG are not able to only amplify C. nobilis but also amplify T. trifolia.
ACKNOWLEDGEMENTS
This study was supported by Udayana University Research Grant No. 383-5/UN
14.4.A/LT//2018
REFERENCES
Badan Pusat Statistik Provinsi Bali. 2018. Produksi buah jeruk dirinci menurut
Kabupaten/Kota di Bali, 2000-2018. https://bali.bps.go.id/dynamictable/ 2017/05/18/128/produksi-ton-buah-jeruk-dirinci-menurut-kabupaten-kota-di-bali-2011-2015.html
Breslauer, K.J., R. Frank, H. Blocker, L. A. Markey. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA, 83:3746-3750.
Jaqoueix, S., J.M Bove, and M. Garnier. 1994. The phloem limited bacterium of greening disease of citrus is as member of the alpha subdivision of proteobacteria. International J. Systemic Bacteriol. 44:397-86.
Kamel, A. 2003. Bioinformatic tools and guideline for PCR primer design. African Journal of Biotechnology 2 (5):91-95.
Mahayani, S. 2013. Analisis Ekspresi Klon Gen CVPDr Dalam Sel Escherichia coli. Jurnal Agroknow 1(1): 33-38.
Martasari, C., A. Supriyanto, Hardiyanto, D. Agisimanto, dan H. Mulyanto. 2004. Keragaman Jeruk Siam di Indonesia. Proseding Seminar Jeruk Siam Nasional. 57-69.
Okonechnikov K, Golosova O, Fursov M, the UGENE team. 2012. Unipro UGENE: a unified bioinformatics
toolkit. Bioinformatics 28: 1166
1167.
Wirawan, I. G. P., S. Liliek dan N. Wijaya. 2004. Penyakit CVPD pada tanaman jeruk. Denpasar. Udayana Press.
Wirawan, I. G. P. 2016. Distribution of CVPDr gene among some citrus plants in bali. International Journal of Bioscience and Biotechnology, 3(2).
Yuniti, I. G. A. D. 2018. Polimorfisme fragmen DNA CVPDr pada beberapa jenis tanaman jeruk di Bali. (Disertasi). Universitas Udayana.
Denpasar.
100 • FACULTY OF AGRICULTURE, UDAYANA UNIVERSITY
Discussion and feedback