PRICK AND SOAK Agroacterium tumefaciens-MEDIATED IN PLANTA TRANSFORMATION IN TOMATO (Lycopersicon esculentum Mill.)
on
INTERNATIONAL JOURNAL OF BIOSCIENCES AND BIOTECHNOLOGY • Vol. 5 No. 2 • April 2018
ISSN: 2303-3371
https://doi.org/10.24843/IJBB.2018.v05.i02.p05
PRICK AND SOAK Agroacterium tumefaciens-MEDIATED IN PLANTA TRANSFORMATION IN TOMATO (Lycopersicon esculentum Mill.)
I Putu Wahyu Sanjaya1*, Rindang Dwiyani2, I G. P. Wirawan2, and Bambang Sugiharto3
1Department Agroecotechnology, Faculty of Agriculture, Udayana University 2Department Agroecotechnology, Faculty of Agriculture, Udayana University, 3Center for Development Advanced Science and Technology, Jember University *Corresponding author: putuw87@ymail.com
ABSTRACT
One of the modern plant breedings through genetic engineering is Agrobacterium tumefaciens-mediated transformation. Agrobacterium tumefaciens-mediated transformation can be performed in vitro or in planta. In planta transformation arises from the weaknesses of the in vitro method such as need high hygiene standard, professional tissue culture experts, and more time to prepare explants and somaclonal variation. In planta transformation is a method to transfer the gene to the plant genome without any tissue culture stages. The aims of this research were to know the possibility of the prick and soak in planta method with the target of tomato seeds and to know the most suitable inoculation time for tomato seeds transformation by prick and soak method the transformation is done by pricking the seeds and soaking them in the A. tumefaciens suspension. The treatments in this study were 1 and 2 days inoculation time to test the efficacy of prick and soak in planta transformation method. Tomato seeds were pricked with a needle on the center once, and then soaked in A. tumefaciens strain LB4404 suspension carrying pKYS-SoSPS1 plasmid with Neomycin Phosphotransferase (NPTII) and Saccharum officinarum Sucrose Phosphate synthase (SoSPS1) genes. Visualization of tomato’s DNA samples after PCR showed that 1-day inoculation sample was positively integrated with NPTII gene and negative in the 2 days inoculation treatment.
Keywords: Tomato seeds, In planta Transformation, Agrobacterium tumefaciens, prick and soak
INTRODUCTION
Tomato is one of the many cultivated plants in Indonesia used as processed materials and fresh consumption. The need for tomatoes requires not only high production but also good quality. Good quality of tomato can be achieved by making tomato plants varieties with superior properties such as high production, response to fertilization, high vitamin content, sweet
taste, fast fruiting, resistant to biotic and abiotic stresses and many more. Tomato plants varieties with superior properties can be obtained by conventional cross-breeding or genetic engineering. Genetic engineering is widely done by scientist lately. Despite its low success rate (Pardal, 2002, Utomo, 2004), genetic engineering is relatively fast (Dwiyani et al., 2016a), can be done with standard laboratory types of equipment
(Rahmawati, 2006) and does not require spacious place compared to conventional cross-breeding (Azrai, 2005).
Genetic engineering in plants can be done by microprojectile bombardment, electroporation, silicon carbide-mediated transformation, and Agrobacterium tumefaciens-mediated transformation (Dwiyani et al., 2016a). A. tumefaciens-mediated transformation is one of the most widely developed genetic engineering methods. A. tumefaciens-mediated transformation method is relatively easy to do, can be done with standard laboratory types of equipment, cheaper cost and plant genome can be derived to the progenies compared to the other genetic engineering methods (Aldemita and Hodges, 1996).
A. tumefaciens-mediated transformation can be done in vitro and in planta. In vitro transformation uses tissue culture stages for transfer gene process, so it requires sterile conditions during the transformation process. In addition, in vitro transformation has several disadvantages including the needs for tissue culture experts, a long time in the provision of explants and sometimes produces somaclonal variation plants (Ping et al., 2003). From the weaknesses in in vitro transformation, the researchers turned to transformation without using any tissue culture stages. The transformation method performed directly without going through the tissue culture
stages is defined as in planta transformation by Feldman and Mark (1987).
In planta transformation has been performed in several plants such as Arabidopsis thaliana (Clough and Bent, 1998; Bent. 2000; Barik, 2013; Narusaka et al., 2010; Zhang et al., 2006), Brassica napus (Li et al., 2010 ), orchids (Semiarti, 2012), Kalanchoe sp. (Dewanto and Suhandono, 2016), wheat (Razzaq et al., 2011), rice (Rod-in et al., 2014; Ratanasut et al., 2017), soybean (Shou et al., 2002; Zia et al., 2011) and tomatoes (Hasan et al., 2008). The floral dip is the most frequent in planta transformation method performed in tomato plants. Floral dip method uses flowers as the target of transformation.
Floral dip in planta transformation stages relatively takes a long time. After transforming the flowers, we still have to wait for the flowers to become ripe fruits, and then the seeds from the fruits grown to be selected as candidate transformants. The idea arises to cut the time of the research by performing a direct transformation on the tomato seed. Until now, there has been no report on the use of tomato seeds directly in in planta transformation. In addition, to shorten the time, the target of the transformation of seeds is also easier to obtain because it is always available in agricultural shops.
This research used in planta transformation method with the target of
tomato seeds. The seeds are treated by pricking and then soaking in the A. tumefaciens suspension so that this method is called the prick and soak method. This method has not been reported by other researchers so the result of this study has novelty value.
According to Dwiyani et al. (2016b) inoculation time becomes an important factor in transformation. This study used soak time as the treatment. Soaking is done for as long as possible with the consideration that the longer the inoculation time, the longer the chances of A. tumefaciens in transferring the gene to the embryo in the seeds. To determine the treatment in this research, there was preliminary research by soaking the pricked seeds for 1 day and 2 days using water. The results showed that the seeds can still germinate after soaked for 2 days with the number of seeds germinate as much as 22%. So it was decided for the treatment in this study is soaking the pricked tomato seeds for 1 and 2 days, because if the seeds soaked longer, it is feared it will make the seeds rotted.
MATERIALS AND METHODS Confirmation of NPTII gene in A.
tumefaciens via PCR (polymerase chain reaction) and electrophoresis
https://doi.org/10.24843/IJBB.2018.v05.i02.p05
The bacteria were strike on a solid LB (Luria Bertani) medium with 50 ppm kanamycin, 100 ppm rifampicin and 12.5 ppm gentamicin antibiotics, incubated in 25ºC for 48 hours. Ten single colonies of A. tumefaciens were taken for amplification on PCR and struck on different points in solid LB medium and marked.Samples were amplified in PCR machine with NPTII (neomycin phosphotransferase) primer ( forward: 5’-
GTCATCTCACCTTCCTCCTGCC 3’;
reverse:
5’GTCGCTTGGTCGGTCATTTCG-3’), NPTII is a kanamycin resistance gene which is the selectable marker in T-DNA construct of the pKYS-SoSPS1 plasmid. NPTII PCR program with predenaturation 95ºC for 3 minutes, denaturation 95ºC for 30 seconds, annealing 59ºC 30 for seconds, extension 72ºC for 1 minute, final extension 72ºC for 5 minutes (Manders et al., 1994). The PCR cycle was performed 35 times with expected target of 550 bp DNA amplified. The results of PCR were electrophoresed then the results were visualized. Positive A. tumefaciens samples containing the NPTII gene were used for transformation.
LB RB
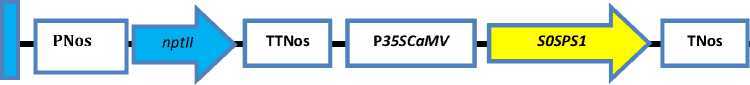
Fig. 1.Construct of T-DNA in pKYS-SoSPS1 plasmid by Sugiharto et al. (1997).
T-DNA in pKYS-SoSPS1 plasmid contain left border (LB), promoter nopaline synthase (PNos), neomycin phosphotransferase (NPTII), terminator nopaline synthase (TNos), promoter CaMV (P35SCaMV), Saccharum officinarum sucrose phosphate synthase (SoSPS1), terminator nopaline synthase (TNos) and right border (RB) as the border of T-DNA
Preparation of A. tumefaciens suspension
PCR positive of single colonies of A. tumefaciens were grown on 2 ml LB medium with 50 ppm kanamycin, 100 ppm rifampicin and 12.5 ppm gentamicin antibiotics then incubated at 150 rpm shaker at 28° C for 24 hours until OD reaches 0.6. Every 1 ml of A. tumefaciens culture was subcultured to 20 ml LB and added with as previous antibiotics concentration then incubated at 150 rpm shaker at 28 ° C for 24 hours until OD reached 0.6. The bacteria are then transferred to the test tubes with a size of 10 ml each tube. The tube is closed and centrifuged at 5,000 rpm for 10 minutes. The supernatant was removed and the precipitate suspended on ½ NP medium containing 100 ppm acetosyringone and 30 µl tween20 until the volume reached 20 ml (Dewanti et al., 2011).
Prick and soak seed transformation
200 tomato seeds were pricked with a needle. Tomato seeds are pricked in the center and then soaked in A. tumefaciens
suspension prepared before. 100 seeds were soaked for one day and the remaining 100 seeds were soaked for two days. The seeds are then filtered and planted in wet tissue on the petri dish to germinate for nine days and then the sprouts transferred in soil medium.
Percentage of seeds that grew after the transformation
After prick and soak transformation
treatment, the germinated seeds were
percentage (GP) with the formula:
GP=
germinated seeds Total seeds transformed
x 100%
Isolation of tomato plant DNA and electrophoresis
Young leaves of the Tomato plants were taken about 3 leaves per plant. The leaves then were put into the plastic and labeled and then stored in ice box containing ice packs. Tomato plant DNA was isolated by a modification of Mini Prep-Method by Zang and Stewart (2000). Modifications include the use of fewer samples and manual grinding method with just a drill bit without the electric drill. The isolated DNA was then
electrophoresed and visualized on the UV cabinet.
PCR and electrophoresis of transformant candidates of tomato plants
Isolated DNA were amplified in PCR machine with NPTII (neomycin
phosphotransferase) primer (forward: 5’-GTCATCTCACCTTCCTCCTGCC-3’; reverse: 5’
GTCGCTTGGTCGGTCATTTCG-3’), NPTII is a kanamycin resistance gene which is the selectable marker in T-DNA construct of the pKYS-SoSPS1 plasmid. NPTII PCR program with predenaturation 95ºC for 3 minutes, denaturation 95ºC for 30 seconds, annealing 59ºC 30 for seconds, extension 72ºC for 1 minute, final extension 72ºC for 5 minutes (Manders et al., 1994). The PCR cycle was performed 35 times with expected target of 550 bp DNA amplified. The result of PCR was electrophoresed then the result
RESULTS AND DISCUSSION NPTII gene confirmation in A.
tumefaciens
Electrophoresis was performed using agarose. The pKYS-SoSPS1 plasmid used as the control. The pKYS-SoSPS1 plasmid contains SoSPS1 gen to increase the production of plant sucrose (Ningtyas et al., 2015) and the NPTII gen as kanamycin resistance gene (Ghanem, 2011). The results of the gene confirmation through PCR showed that the tenth single colony samples from A. tumefaciens grown on solid LB medium containing the NPTII gene which can be seen in Fig. 2. It appears that the tenth single colonies of A. tumefaciens amplified positive containing pKYS-SoSPS1 plasmid, so it can be ascertained that the tenth single colonies of A. tumefaciens used for the transformation in this research positively contain the SoSPS1 and NPTII genes.
was visualized.
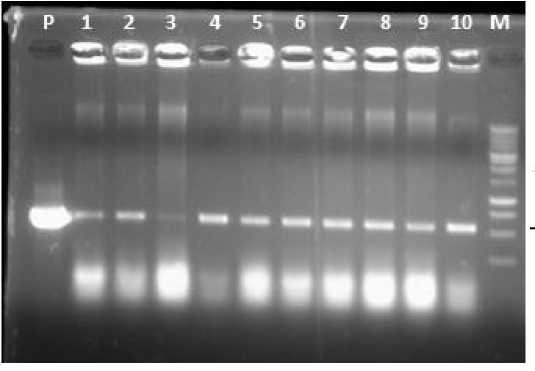
Fig. 2. The gene confirmation in the A. tumefaciens using NPTII primers.
P = pKYS-SoSPS1 plasmid; 1-10=single colonies of A. tumefaciens; M=DNA marker of 1 kb DNA ladder.
♦ 550 bp
Percentage of seeds germinated
Seeds that germinated after pricked and soaked were 22% in the 1-day soaking treatment and 18% in 2 days soaking treatment. Germinated seeds after transformed were planted in the soil until 13
days (Fig. 3). The low number of seeds germination caused by the seeds were pricked, this can be concluded that the tomato seeds used in this research has good quality because there were 98% of seeds germinated in control treatment or without pricked.

Fig. 3. Tomato seeds Germination and grow after transformed.
A = Seed germinates after 1-day soaking treatment;
B = Seed germinate after 2-days soaking treatment;
C = Seeds which become plants after the transformation of 1-day soaking treatment;
D = Seeds that become plants after the transformation of 2-days soaking treatment.
Pricking of tomato seeds in the middle with the target endosperm was done to injure and penetrate the seed coat. The wound on the seeds provides access for A. tumefaciens into the seeds to infecting the embryo in tomato seeds. The low percentage of the germinated seeds due to the embryo or the endosperm damage. Pricking tomato seeds with a needle may be damaged the embryo so that the seeds are not able to germinate.
According to Syukur et al. (2012), the embryo in the seed is a tiny plant that will germinate and grow into a plant, so that if the embryo on the seed were damaged it will interfere the seed germination. The growth of the sprouts becomes slow and thin because the prick treatment damages the endosperm. Endosperm in the seed intends as a food or nutrient supplier to maintain the embryo during the process of embryo development
into sprouts (Sukamto, 2011). Damage to the endosperm in the seed affects the number of nutrients absorbed by the seed for the early development of tomato embryos.
Tomato plant DNA isolation
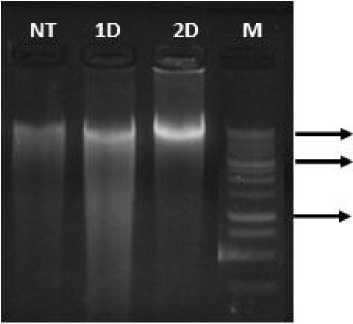
IOOOO bp 3000 bp
1000 bp
Fig. 4. Visualization of DNA isolation result from tomato leaves samples.
NT = non-transformant sample; 1D = 1 day immersion; 2D = 2 days immersion; and M = DNA marker 1 kb DNA ladder.
Fig. 4 shows that DNA from tomato leaves can be extracted using the modification of Mini-Prep method from
Zang and Stewart (2000). The number of samples used in this research only 0.03 grams per sample from the recommended 0.3 grams per sample in the Mini-Prep method. Thus, this method can be used to extract DNA from the leaves of tomato plants with fewer samples needed in the Mini-Prep method by Zang and Stewart (2000).
Molecular analysis of transformant candidates of tomato plants
The results of this research showed that the prick and soak in planta transformation with the treatment of 1-day soaking of tomato seeds was able to integrate T-DNA into the tomato plants genome. The integration of the NPTII gene with plant genome is also accompanied by the integration of the SoSPS1 gene. The use of NPTII as a primer because of NPTII is a specific gene not owned by tomato plants (Meissner et al., 1997).
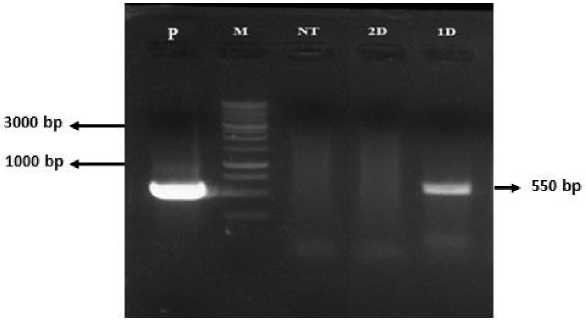
Fig. 5. Electrophoresis visualization of the PCR results from isolated DNA of
transformant candidates of tomato plants using primers of the NPTII gene. P = pKYS-SoSPS1 plasmid, M = DNA marker of 1 kb DNA ladder, NT = non transformed plant, 2D = 2 days soaking treatment, and 1D = 1 day soaking treatment.
In planta transformation has been successfully performed using the floral dip (Ratanasut et al., 2017; Bent 2000; Li et al., 2010; Rod-in et al., 2014; Narusaka et al., 2010; Zang et al., 2006 Clough and Bent 1998), by soaking the flower of the plant in the liquid A. tumefaciens culture; the pin pricking (Razzaq et al., 2011; Jaganath et al., 2013; Seol et al., 2008), by pricking the transformation target with the needle smeared with A. tumefaciens; agro-injection (Bratic et al., 2007; Orzaez et al., 2006; Zia et al., 2011), by injecting A. tumefaciens culture directly into the plant tissue or organs; vacuum infiltration (Nanasato et al., 2013; Bratic et al., 2007), by a vacuum to force A. tumefaciens get in into plant cells; and polen-tube pathway (Semiarti et al., 2012; Shou et al., 2002; Song et al., 2007; Zang and Luan, 2016), by immersing pollen from male flowers with A. tumefaciens culture then breed it with female flowers to produce a transformant progeny. There haven’t been any reports of transformation done by pricked and soaked the seeds as was done in this research.
The prick and soak in planta transformation method is a combination of pricking pin and floral dip methods. The pricking of the transformant target using the needle adopted from the pin pricking method while the soak of the transformation target with the suspension of A. tumefaciens adopted the floral dip method. Pin pricking
method is done by smearing sterile needle with A. tumefaciens culture then the needle is used to pierce the seed or other plants tissue (Seol et al., 2008), while the floral dip method is done by soaking the existing flower on the plant without injuring the flower (Clough and Bent, 1998).
The pin pricking method is considered to be less efficient because the smeared needle has a narrow surface area. This results the number of bacterias carried by the needle are limited, affecting the low chance for transformation. The weakness of the floral dip method lies in the length of time required to obtain the transformant plant. Floral dip method can be done if the plants already have flowers. After transforming the flowers, we still have to wait for the flowers to become fruits to the physiological ripe so that the seeds can be grown to be selected as the candidate transformant plants. The disadvantages of the pin pricking and floral dip methods inspired the prick and soak method. The prick and soak method is performed by prick the transformation target and then soak it with the suspension of A. tumefaciens.
Soaking the seeds that have been pricked once with the A. tumefaciens suspension done with the aim to provide nutrition, optimal environment to facilitate A. tumefaciens to move to the plant cells so that bacterial infections to the plant cells more evenly to all plant cells that submerged have
the possibility to be infected. According to Dewanto and Suhandono (2016), soaking media is important in transformation because the media supports optimal conditions that can promote the interactions and increase gene transfer rates and weaken plant cells defenses to facilitate bacterial infections. It shows that the use of the soaking media has an effect on transformation.
The time of soaking or inoculation time has an effect on the transformation. According to Dwiyani et al. (2016b), inoculation times becomes an important factor in transformation. Soaking for 1 day gives the best result of transformation compared to soaking for 2 days. Allegedly, there is an optimal inoculation or soaking time to obtain a high transformation efficiency, if passing a certain time then the number of transformants obtained will be reduced. Purnamaningsih (2010) in in vitro transformation found that the inoculation treatment of tomatoes cotyledon showed the inoculation time for 45 minutes gave the best result from the treatment of 0, 15, 30 and 45 minutes of A. tumefaciens inoculation. However, in a study conducted by Shrestha et al. (2007) using orchid protocorm in vitro with 10, 60, 240 and 480 minutes inoculation treatments showed that the 240-minute inoculation treatment resulted in the highest number of transformants.
https://doi.org/10.24843/IJBB.2018.v05.i02.p05
The longer interaction between plant cells and A. tumefaciens caused the plant to detect the presence of A. tumefaciens as a pathogen and begin to increase the resistance system and limit the activity of the transformation by hypersensitive reaction (Coll et al., 2011). According to Jones and Dangl (2006), hypersensitive reactions are active reactions when the pathogen interfere with molecular plant cells. The stage begins with the plant recognizing the pathogen that enters and begins to activate the resistance system. Successful pathogens propagate virulent effector. Effector recognized by plant immune systems send signals to nucleotides. Nucleotide response form is resistant to the pathogen by programmed cell death (hypersensitive reaction).
Another thing that supports the success of transformation in this research is the use of tween20 and acetosyringone (AS). Clough and Bent (1998) suggest that the addition of surfactants at low concentrations in inoculation media can increase the number of transformants obtained in Arabidopsis thaliana plants using the floral dip method. Tween20 is one of surfactant that serves to reduce the surface tension of the cell so that hydrophobic reactions of plant cells and bacterial cells decreases and facilitates the attachment of A. tumefaciens in plant cells (Miller, 2013).
Research conducted by Sheikholeslam and Weeks (1987) in Arabidopsis thaliana showed that addition of acetosyringone able to increase the number of transformants up to 63% than without giving acetosyringone which only gets 2-3% transformants. A. tumefaciens is a gram-negative bacteria that carries the transfer DNA (T-DNA) in its plasmid and is able to infect through wounds in the plant. The plasmid of A. tumefaciens is composed of left border, T-DNA, right border, and virulence region (virgen). AS compounds play a role in activating virgen to release T-DNA from plasmids and deliver T-DNA to plant cells to integrate with plant genomes (Afolabi-Balogun, 2014).
ACKNOWLEDGMENT
Author wishes to thank Prof. Dr. Ir. Rindang Dwiyani, M.Sc. for the financial and also as an adviser and also acknowledge the Centre Development of Advanced Sciences and Technology (CDAST), Jember University, for facilitating the molecular research.
REFERENCES
Afolabi-Balogun, N. B., Inuwa, H. M., Ishiyaku, M. F., Bakare-Odunola, M. T., Nok, A. J., & Adebola, P. A.
(2014). Effect of Acetosyringone on Agrobacterium-Mediated Transformation of Cotton. ARPN Journal of Agricultural and Biological Science, 9(8), 284-286.
Aldemita, R. R., & Hodges, T. K. (1996). Agrobacterium tumefaciens-Mediated Transformation of Japonica and Indica Rice Varieties. Planta, 199(4), 612
617.
Azrai, M. (2005). Ulasan Pemanfaatan Markah Molekuler dalam Proses, Pemuliaan Tanaman. AgroBiogen, 1(1), 26 – 37.
Barik, S. K. (2013). Floral Dip: A Simple and Efficient Agrobacterium-Mediated Transformation Method is Used in A Model Plant Arabidopsis thaliana. Cibtech Journal of Bio-Protocols, 2 (2): 14-22.
Bent, A. F. (2000). Arabidopsis in Planta Transformation. Uses, Mechanisms, and Prospects for Transformation of Other Species1. Plant Physiology, 124: 1540-1547.
Bratic, A. M., Majic, D. B., Miljus-Djukic, J. D., Jovanovic, Z. S., & Maksimovic, V. R. (2007). In Planta Transformation of Buckwheat (Fagopyrum esculentum Moench.). Arch. Biol. Sci. Belgrade, 59(2): 135-138.
Clough, S. J., & Bent, F. (1998). Floral Dip: A Simplified Method for
Agrobacterium-Mediated Transformation of Arabidopsis thaliana. The Plant Journal, 16(6),
735–743.
Coll, N. S., Epple, P., & Dangl, J. L. (2011). Programmed Cell Death in The Plant Immune System. Cell Death Differ, 18, 1247–1256.
Dewanti, P., Islahuddin, M., Okviandari, P., Waluyo, S., Saputra, B. A., Wardiyati, T., & Sugiharto, B. (2011). Efisiensi Transformasi Tomat (Lycopersicon esculentum) dengan Gen SoSPS1
Menggunakan Agrobacterium
tumefaciens. Berk. Penel. Hayati Edisi Khusus, 4C: 73–78.
Dewanto, H. A., & Suhandono, S. (2016). Transformasi Menggunakan
Agrobacterium tumefaciens Pada Tunas Daun Kalanchoe mortagei dan Kalanchoe deigremontiana 1 Dan 2. Chimica Et Natura Acta, 4(2): 7-105.
Dwiyani, R., Yuswanti, H., Darmawati, I. A. P., & Mayadewi, N. N. A. (2016a). Transformasi Genetik pada Tanaman Melalui Agrobacterium tumefaciens. Swasta Nulus; Bali.
Dwiyani, R., Yuswanti, H., Mercuriani, I. S., & Semiarti, E. (2016b). Transformasi Gen Pembungaan melalui
Agrobacterium tumefaciens Secara In-Vitro pada Tanaman Anggrek Vanda tricolor. Agrotrop, 6(1), 83 – 89.
Ghanem, S. (2011). Cloning of The NPTII Gene of Escherichia coli and Construction of A Recombinant Strain Harboring Functional recA and NPTII Antibiotic Resistance. Genetics and Molecular Research, 10 (3), 1445-1454.
Hasan, M., Khan, A. J., Khan, S., Shah, A. H., Khan, A. R., & Mirza, B. (2008). Transformation of Tomato
(Lycopersicon esculentum Mill.) with Arabidopsis Early Flowering Gene Apetalai (Api) Through Agrobacterium Infiltration of Ripened Fruits. Pak. J. Bot, 40(1), 161-173.
Jaganath, B., Subramanyam, K., Mayavam, S., Karthik, S., Elayaraja, D., Udayakumar, R., Manickavasagam, M., & Ganapathi, A. (2013). An
Efficient In Planta Transformation of Jatropha curcas (L.) and
Multiplication of Transformed Plants Through In Vivo Grafting.
Protoplasma, 251(3), 591-601.
Jones J. D. & Dangl, J. L. (2006). The Plant Immune System. Nature, 444, 323-329.
Li, j., Tan, X., Zhu, F., & Guo, J. (2010). A Rapid and Simple Method for Brassica Napus Floral-Dip Transformation and Selection of Transgenic Plantlets. International Journal of Biology, 2(1), 127-131.
Manders, G., Otoni, W. C., Vaz, F. B. D. U., Blackhall, N. W., Power, J. B., &
Davey, M. R. (1994). Transformation of Passionfruit (Passiflora edulis fv flavicarpa Degener.) using
Agrobacterium tumefaciens. Plant Cell Report, 13, 697-702.
Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., Elkind, Y., & Levy, A. (1997). A New Model System for Tomato Genetics. The Plants Journal, 12(6), 1465-1472.
Miller, P. D. (2013). Method for Improved Transformation Using Agrobacterium. United States Patent Application Publication. US2013/0157369A1: 1-14.
Nanasato, Y., Okuzaki, A., & Tabei, Y. (2013). Improving The Transformation Efficiency of Cucurbita Species: Factors and Strategy for Practical Application. Plant Biotechnology, 30, 287-294.
Narusaka, M., Shiraishi, T., Iwabuchi, M., & Narusaka, Y. (2010). The Floral Inoculating Protocol: A Simplified Arabidopsis thaliana Transformation Method Modified from Floral Dipping. Plant Biotechnology, 27, 349-351.
Ningtyas, R. M., Sugiharto, B., & Utarti, E. (2015). Transformasi Gen Sosps1 pada Tanaman Tebu Overekspresi Gen Sosut1 Event 2 Menggunakan Agrobacterium Tumefaciens. Berkala sainstek, 3(1), 20-23.
Orzaez, D., Mirabel, S., Wieland, W. H., & Granell, A. (2006). Agroinjection of Tomato Fruits. A Tool for Rapid Functional Analysis of Transgenes Directly in Fruit1. Plant Physiology, 140, 3-11.
Pardal, S. J. (2002). Perkembangan Penelitian Regenerasi dan
Transformasi pada Tanaman Kedelai. Buletin AgroBio, 5(2), 37 – 44.
Ping, L. X., Nogawa, M., Shioiri, H., Nozue, M., Makita, N., Takeda, M., Bao, L., & Kojima, M. (2003). In Planta Transformation of Mulberry Trees (Morus alba L.) by Agrobacterium tumefaciens. Journal of Insect Biotechnology and Sericology, 72,
177-184.
Purnamaningsih, R. (2010). Introduksi Gen DefH9-iaaM dan DefH9-RI-iaaM ke dalam Genom Tanaman Tomat
Menggunakan Vektor Agrobacterium tumefaciens. AgroBiogen, 6(1), 18-25.
Rahmawati, S. (2006). Status Perkembangan Perbaikan Sifat Genetik Padi Menggunakan Transformasi
Agrobacterium. AgroBiogen, 2(1), 36 – 44.
Ratanasut, K., Rod-In, W., & Sujipuli, K. (2017). In Planta Agrobacterium-
Mediated Transformation of Rice. Science Direct, 24(3), 181-186.
Razzaq, A., Hafiz, I. A., Mahmood, I., & Hussain, A. (2011). Development of In Planta Transformation Protocol for Wheat. African Journal of Biotechnology, 10(5), 740-750.
Rod-in, W., Sujipuli, K., & Ratanasut, K. (2014). The Floral-Dip Method for Rice (Oryza sativa) Transformation. Journal of Agricultural Technology, 10(2), 467-474.
Semiarti, E., Purwantoro, A., Mercuriani, I. S., Anggriasari, A. M., Jang, S., Machida, Y., & Machida, C. (2012). In Planta Transformation Method for T-DNA Transfer In Orchids. International Conference on
Mathematics and Natural Sciences, 1589, 303-307.
Seol, E., Jung, Y., Lee, J., Cho, C., Kim, T., Rhee, Y., & Lee, S. (2008). In Planta Transformation of Notocactus scopa cv. Soonjung By Agrobacterium tumefaciens. Plant Cell Reports, 27(7), 1197-1206.
Sheikholeslam, S. N., & Weeks, D. P.
(1986). Acetosyringone Promotes High-Efficiency Transformation of Arabidopsis thaliana Explants
by Agrobacterium tumefaciens. Plant Molecular Biology, 8(4), 291-298.
Shou, H., Palmer, R. G., & Wang, K. (2002). Irreproducibility of the Soybean Pollen-Tube Pathway Transformation Procedure. Plant Molecular Biology Reporter, 20, 325–334.
Shrestha, B. R., Chin, D. P., Tokuhara, K., & Mii, M. (2007). Efficient Production of Transgenic Plants of Vanda through Sonication-Assisted Agrobacterium-
Mediated Transformation of
Protocorm-Like Bodies. Plant
Biotechnology, 24: 429-434.
Song, X., Gu, Y., & Qin, G. (2007).
Application of A Transformation Method Via The Pollen-Tube Pathway In Agriculture Molecular Breeding. Life Science Journal, 4(1), 77-7.
Sugiharto, B., Sakakibara, Sumadi, & Sugiyama. (1997). Differential
Expression of Two Genes for SucrosePhosphate Synthase in Sugarcane: Molecular Cloning of the cDNAs and Comparative Analysis of Gene Expression. J. Plant Cell Physiol, 38, 961-965.
Sukamto, L. A. (2011). Induksi Tanaman Cendana (Santalum album L.) Triploid Melalui Kultur Endosperma Secara In Vitro.Widyariset, 4(2), 393-398.
Syukur, M., Sujiprihati, S., & Yunianti, R. (2012). Teknik Pemuliaan Tanaman. Penebar Swadaya : Jakarta.
Utomo, S. D. (2004). Pengaruh Strain Agrobacterium Terhadap Efisiensi Transformasi Genetik Jagung Genotipe Hibrida Hiii. Ilmu Pertanian, 11(2), 1 – 11.
Zang, H. J., & Luan, F. S. (2016).
Transformation of The CmACS-7
Gene Into Melon (Cucumis melo L.) Using The Pollen-Tube Pathway. Genetics and Molecular Research, 15 (3), 1-10.
Zang, J., & Stewart, J. McD. (2000).
Economical and Rapid Method for Extracting Cotton Genomic DNA. The Journal of Cotton Science, 4: 193-201.
Zang, X., Henriques, R., Lin, S., Niu, Q., & Chua, N. (2006). Agrobacterium-Mediated Transformation of
Arabidopsis thaliana Using The Floral Dip Method. Nature Protocols, 1(2), 16.
Zia, M., Arshad, W., Bibi, Y., Nisa, S., & Chaudhary, M. F. (2011). Does AgroInjection To Soybean Pods Transform Embryos?. Plant Omics Journal, 4(7):384-390.
135• ASIA OCEANIA BIOSCIENCE AND BIOTECHNOLOGY
Discussion and feedback