GENETIC DIVERSITY OF GUAVA (Psidium Guajava L.) IN BALI INDONESIA BASED ON RAPD MARKER
on
ORIGINAL RESEARCH ARTICLE
GENETIC DIVERSITY OF GUAVA (Psidium Guajava L.) IN BALI INDONESIA BASED ON RAPD MARKER
Ni Nyoman Ari Mayadewi1*, I Nyoman Rai1,, Rindang Dwiyani1, and Ida Ayu Astarini2
1Department of Agroecotechnology, Faculty of Agriculture, Udayana University, Bali 2Department of Biology, Faculty of Mathematics and Natural Sciences,
Udayana University, Bali
* Corresponding author : arimayadewi@unud.ac.id
Received : 7th June 2016 | Accepted : 22nd July 2016
ABSTRACT
The aim of this study was to investigate genetic diversity and relationship of guava (local name: jambu biji) genotypes grown in Bali, Indonesia, based on Random Amplified Polymorphic DNA (RAPD) markers. Twelve guava cultivars namely Jambu biji ‘Australia’, ‘Bangkok Merah’, ‘Bangkok Putih’, ‘Cokorde’, ‘Dadu 1’, ‘Dadu 2’, ‘Getas Merah’, ‘Kamboja’, ‘Kristal’, ‘Pipit’, ‘Susu’ and ‘Variegata’ were collected from nine Regencies in Bali using survey and explorative method. Ten decamers RAPD primers were employed to distinguish between the 12 cultivars and determine their genetic relationships. A dendogram was constructed using coefficient dissimilarity analysis based on phylogenetic analysis using parsimony (PAUP). The twelve cultivars were grouped into 2 main clusters and five smaller clusters. Variation between genotypes of guava local will be good sources for future crop improvemen.
Keywords: DNA markers, RAPD analysis, guava cultivars
INTRODUCTION
Guava (Psidium guajava L, family Myrtaceae) is one of the 50 most well-known edible fruit (Chandra & Mishra, 2007) and are commonly cultivated in tropical region including Indonesia. Guava is an allogamous and highly heterozygous fruit crop. Many variations occur in local Indonesia guava as a result of traditional seedling selection by local people. Vegetative propagation was done rarely, so that variations within cultivars are quiet high. In Indonesia, guava mainly
grows as backyard trees, not an intensive farming. Guava can be consumed fresh as table fruit, or processed in guava juice, jelly, jam and sweets.
Demand for guava in Indonesia is high, both for fresh fruit and processing into juice. Guava fruit is rich in dietary fiber, contain high vitamin A, C, calcium, potassium and iron (Valdes et al. 2007). Guava also high on quercetin, which is effective to increase blood thrombocyte and therefore the fruit juice is a common
ISSN ONLINE: 9 772303 337 008
supplement for dengue fever patients in Indonesia. In addition, guava leaves extract contain tannin which is effective to treat acute diarrhea and gastroenteritis (Soedarya 2010) and are commonly use in Balinese community.
Diversity of local guava in Indonesia needs to be investigated to be able to select favorable and superior genotypes for further cultivation and improvement. Morphological traits such as leaf shape, leaf size, fruit size and fruit color are important phenotypic markers to select superior genotypes. However, morphological traits may change with the agronomic treatment and growth environment. Therefore, molecular characterization needs to be developed to distinguish between genotypes. RAPD marker has been used to distinguish guava genotypes in India (Chandra and Mishra, 2007), Bangladesh (Ahmed et al., 2011), Taiwan (Chen et al., 2007) and Kenya (Kidaha et al., 2014). RAPD marker has also been employed on wani, a native fruit of Bali (Rai et al., 2008) and cauliflower (Astarini et al., 2002; Astarini et al., 2006). In this study, RAPD analysis will be employed to distinguish guava genotypes that are grown in Bali,
Indonesia.
The use of RAPD technique on perennial plants would increase selection efficiency since it can be done on young plants. RAPD data will complement morphological data and has a great potential on developing germplasms collection data base (Rai et al., 2008).
MATERIALS AND METHODS Study site and sampling
Guava samples were taken from 9 Regencies of Bali, Indonesia, namely Badung, Bangli, Buleleng, Tabanan, Gianyar, Klungkung, Karangasem, Jembrana and Denpasar. Sites altitude ranging from 200 – 700 m above sea level, and temperature ranging from 20 – 30oC. Purposive sampling technique (Rai et al., 2008) was employed and selected trees were tagged for study.
RAPD analysis
DNA extraction
Guava young leaves were collected from field and placed in plastic bag containing silica gel. Leaves were kept in ice box until arriving in lab and then keep in -20oC freezer until use. Guava leaves were washed under running tap water and then dried. Guava leaf, 0.1 gram, was
weighed and crushed using mortar and pestle, then 800 µl of extraction buffer (CTAB, consists of 2% CTAB, 1.4 M NaCl, 100 mM Tris HCl pH 8, 20 mM EDTA pH 8, 1% β-mercaptoethanol
that has been incubated on 65oC waterbath for 30 minute) was added. Sample mixture was then incubated on 65oC waterbath for 60 minute, and shake gently every 10 minutes during incubation. Samples were then
centrifuged for 15 mins at 12,000 rpm and the supernatant were mixed with equal volumes of chloroform: isoamyl alcohol (24:1). The mixture was centrifuged at 12,000 rpm for 15 mins. Supernatant were taken gently and mixed with an equal volume of cold isopropanol and incubated at -20oC for at least 1 hour. The nucleic acid was pelleted by centrifugation for 10 minute at 12,000 rpm and the pellet washed with 500 µl of 70% ethanol and then centrifuged for 5 minute at 12,000 rpm. The DNA pellet was air dried and resuspended in 50 -100 µl TE buffer + 1% RNAse on 37oC waterbath for 60 minute. Purified DNA was then stored at 4oC refrigerator until use.
DNA Quantification
Purified DNA was quantified using Gene Quant to find out DNA
concentration obtained. Reference was set using psdH2O. Dilution factor (psd H20) approximately 1998 µl and DNA about 2 µl was entered into cuvette and then entered into Gene Quant to measure light absorption at 260 nm wavelength so DNA concentration and DNA-RNA ratio can be determined.
DNA dilution.
DNA dilution was done with the aim to obtain DNA concentration according to amplification requirement, i.e 2.5 ng/µl. DNA solution was added with psdH2O until required DNA was achieved.
DNA amplification using PCR machine
Ten RAPD primers suitable for guava were used for DNA amplification. Primers name and nucleotide sequences were OPA 11 (5’ CAATCGCCGT 3’), OPA 20 (5’ GTTGCGATCC 3’), OPB 7 (5’ GGTGACGCAG 3’), OPB 8 (5’ GTCCACACGG 3’), OPB 10 (5’ CTGCTGGGAC 3’), OPB 18 (5’ CCACAGCAGT 3’), OPC 8 (5’ TGGACCGGTG 3’), OPD 5 (5’ TGAGCGGACA 3’), OPD 11 (5’ AGCGCCATTG 3’), OPD 20 (5’ ACCCGGTCAC 3’). Primers were synthesis by Life Technologies Inc.
ISSN ONLINE: 9 772303 337 008
(Madison) customer primer program.
µl total volumes, comprised of 5 µl
The PCR reaction was performed in 10 0.25 µl 100 µM primer (Sigma-Prolgo), 2.5 µl DNA sample (template) and 2.25 sterile aquabidest. DNA
PCR mix Go Taq® Green (Promega), were visualized under UV light to obtained polymorphic bands. Data was obtained by scoring polymorphic band
amplification was done using BOECO
of each genotype on each size. Band
PCR System. Initial hot start was done at 95oC for 1 min, followed by 45 cycles of denaturation at 95oC for 45 second, annealing at 37oC for 1 min, elongation at 72oC for 1 min and 30
size was estimated by comparing with 100 bp ladder (VC 100 bp Plus DNA ladder). Bands were scored as 0 for absent and 1 for present. Genetic distance and phylogenic analysis were
second, followed with a final extension
done using software package Genalex
at 72oC for 7 min.
6.1 and Ntsys pc 2.2.
Gel electrophoresis and data analysis.
Amplified DNA was run on 1% (b/v) agarose that has been added with ethidium bromide for staining, on 1X TBE (consists of 0.45 M Tris-HCl pH 8, 0.45 M Boric acid, 20 mM EDTA).
RESULTS AND DISCUSSION
All 10 RAPD primers produced multiple PCR fragments. Total 118 polymorphics bands were scored. The molecular weight of bands amplified from ten primers ranged from 150 bp to
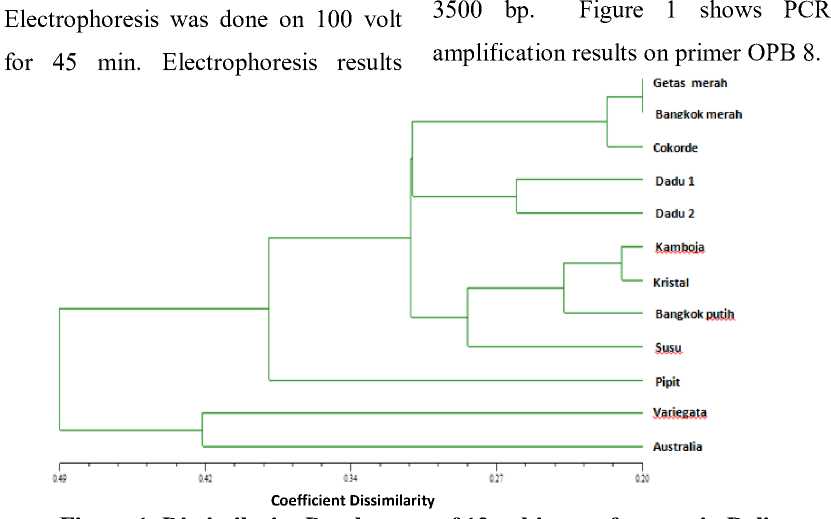
Figure 1 Dissimilarity Dendogram of 12 cultivars of guava in Bali.
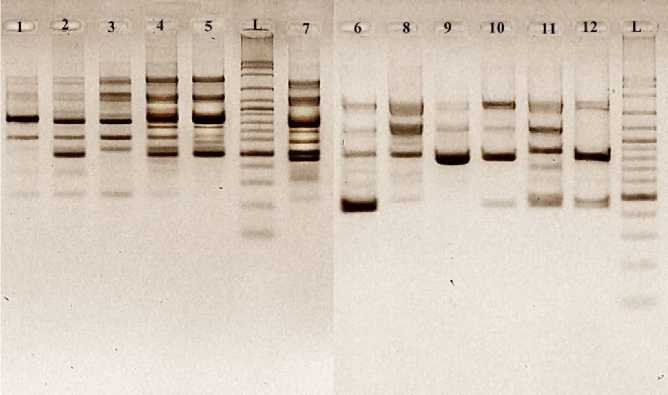
Figure 2 Amplification results using Primer OPB 8.
A dendogram showing relationship among 12 cultivars was generated (Fig. 2). Two major clusters were obtained. First cluster consists of ‘Variegata’ and ‘Australia’ cultivars, which were on separate cluster with 10 other cultivars, with 0.49 coefficient dissimilarity. ‘Variegata’ and ‘Australia’ cultivars have a similar fruit shape (round), red flesh and spreaded seed. Although their fruits are very similar, cultivars of ‘Variegata’ and ‘Australia’ have different flower colour, i.e. white for ‘Variegata’ and red for ‘Australia. Leaf shapes are similar but ‘Variegata’ has a combination of white and green on its leaf, while ‘Australia’ has solid green colour. Cultivar of ‘Australia’ in particular, has specific characteristics, i.e. its stem and fruit has purplish red colour. Fruit productivity is very low,
and therefore this cultivar often use as ornamental fruit plants in pot. Cultivar
of ‘Variegata’ also often us as ornamental pot plants as it has colorful leaf, produce a lot of fruit but plain taste.
Although cultivar ‘Pipit’ was in one big cluster with the 9 other cultivars, it has a quite distant relationship with the other, as revealed by their dissimilarity coefficient of 0.39. ‘Pipit’ has distinct characteristics; smallest fruit compared to others and has white flesh with yellow colour thin skin. Ripe fruits have a beautiful fragrance and sweet taste.
Cultivars of ‘Getas Merah and ‘Bangkok Merah’ are very closely related, as revealed by their coefficient dissimilarity of 0.2. Name ‘Bangkok’ means the plant was originally from Bangkok, Thailand. ‘Getas Merah’ is a
ISSN ONLINE: 9 772303 337 008
result of a cross between ‘Bangkok Merah’ with local guava from Pasar Minggu, Jakarta. ‘Getas Merah’ has many good fruit characteristics including bright red flesh, thick flesh, sweet taste with nice fragrance. The plant has a strong rooting system and highly responsive to fertilizer, and good level of pest and disease resistance, so that it has high fruit productivity
Cultivars of ‘Dadu 1’ and ‘Dadu 2’ were in the same cluster with coefficient dissimilarity about 0.26. Both genotypes have pale pink flesh, thick flesh and small number of seed. The fruit shape was a bit different,
where ‘Dadu 2’ has a round shape, while ‘Dadu 1 has narrower fruit shape toward the base of the fruit.
Another group are cultivars of ‘Kamboja’, ‘Kristal’, ‘Bangkok Putih’ and ‘Susu’. All cultivars have white and thick flesh with low seed number. Cultivars of ‘Bangkok Putih’ has a crunchy texture and sweet taste. It has light green skin colour when ripe. Cultivar of ‘Susu’ has a soft flesh and therefore will be a perfect choice for processing into jam, juice or jelly.This study demonstrated that RAPD analysis provide a simple and reliable method for cultivar identification.
Tabel 1 Genetic dissimilarity matrix between 12 cultivars of guava in Bali.
|
Kultivar |
Getas Merah |
Bangkok Merah |
Varie-gata |
Cokor-de |
Dadu 1 |
Australia |
Pipit |
Kamboja |
Kristal |
Dadu 2 |
Bangkok Putih |
Susu |
|
Getas Merah |
0 | |||||||||||
|
Bangkok Merah |
24 |
0 | ||||||||||
|
Variegata |
44 |
46 |
0 | |||||||||
|
Cokorde |
29 |
27 |
51 |
0 | ||||||||
|
Dadu 1 |
41 |
35 |
57 |
36 |
0 | |||||||
|
Australia |
46 |
40 |
40 |
45 |
37 |
0 | ||||||
|
Pipit |
41 |
35 |
45 |
38 |
44 |
51 |
0 | |||||
|
Kamboja |
31 |
31 |
55 |
40 |
36 |
53 |
46 |
0 | ||||
|
Kristal |
35 |
31 |
49 |
34 |
36 |
49 |
40 |
24 |
0 | |||
|
Dadu 2 |
40 |
34 |
54 |
35 |
31 |
40 |
47 |
35 |
35 |
0 | ||
|
Bangkok Putih |
43 |
29 |
53 |
42 |
32 |
47 |
46 |
28 |
28 |
29 |
0 | |
|
Susu |
38 |
40 |
56 |
41 |
43 |
50 |
41 |
25 |
35 |
38 |
35 |
0 |
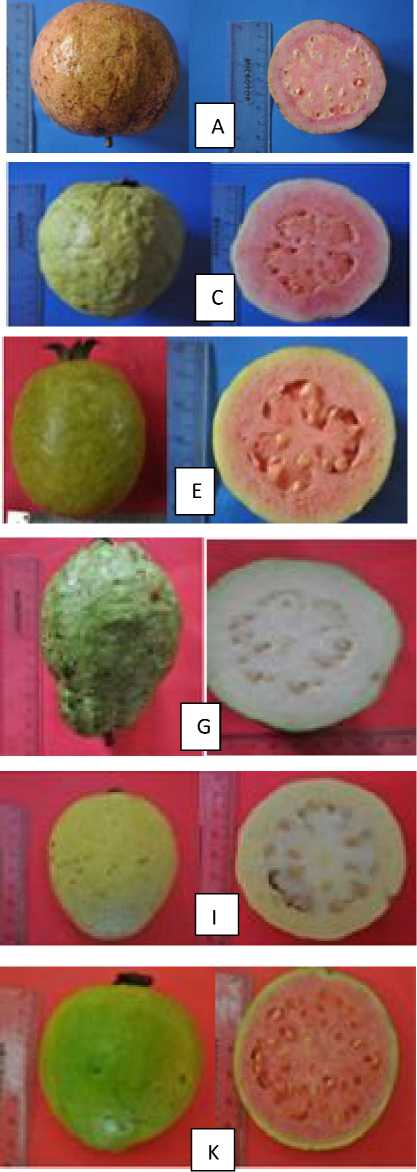
Figure 3 Guava diversity in Bali. a) Australia, b) Bangkok putih, c) Cokorde, d) Dadu 1, e) Dadu 2, f) Getas Merah, g) Kamboja, h) Kristal, i) Pipit, j) Susu, k) Variegata, l) Bangkok merah.
ISSN ONLINE: 9 772303 337 008
ACKNOWLEDGEMENTS
We thank The Ministry of Research, Technology and Higher Education of Indonesia for providing Doctoral scholarship to Ni Nyoman Ari Mayadewi.
REFERENCES
Ahmed, B., M. A. Mannan and S. A.
Hossain. 2011. Molecular characterization of guava (Psidium guajava L.) germplasm by RAPD analysis. Intl. J. Nat. Sci. 1:62-67.
Astarini, I. A., J. A. Plummer, R. A. Lancaster, G. Yan. 2004.
Fingerprinting of cauliflower
cultivars using RAPD markers.
Aust. J. Agric. Res. 55:117–124.
Astarini, I. A., J. A. Plummer, R. A. Lancaster, G. Yan. 2006. Genetic diversity of Indonesian cauliflower cultivars and their relationships with hybrid cultivars grown in Australia. Sci. Hort. 108: 143–150.
Chandra R. and M. Mishra. 2007. Biotechnological Interventions for Improvement of Guava (Psidium guajava L.). Acta Hort. 735:117126.
Chen T.W., C. C. Ng, C. Y. Wang and Y. T. Shyu. 2007. Molecular
Identification and Analysis of Psidium guajava L. from Indigenous Tribes of Taiwan. J. Food and Drug Analysis. 1:82-88.
Kidaha, L. M., A. E. Alakonya, A. B.
Nyende. 2014. Molecular
Characterization of Guava
Landraces in Kenya (Western and South Coast). J. Biol. Agric. and Healthcare Journal of Biology, Agriculture and Healthcare 4 (15): 81-87.
Rai, I.N., G. Wijana, and C. G. A. Semarajaya. 2008. Identifikasi Variabilitas Genetik Wani Bali (Mangifera caesia Jack.) dengan Analisis Penanda RAPD. J. Hort. 18 (2):125-134
Soedarya, A.P. 2010. Agribisnis Guava (Jambu Batu). Budidaya-Usaha-Pengolahan. Penerbit CV. Pustaka Grafika. Bandung. pp188.
Valdes, I. J., D. Becker, N. Rodriguez, B. Velazquez, G. Gonzalez, D. Sourd, L. Rodriguez, E. Ritter, W. Rohde. 2003. Molecular
characterization of Cuban
genotypes of guava, establishment of a first molecular linkage map and mapping of QTLs for vegetative characters. J. Genet. & Breed. 57: 349-358.
8 • ASIA OCEANIA BIOSCIENCES AND BIOTECHNOLOGY CONSORTIUM
Discussion and feedback