Identification of Viral Nervous Necrosis (VNN) in Grouper Fish (Epinephelus sp.) by Reverse Transcriptase-Polymerase Chain Reaction
on

Advances in Tropical Biodiversity and
Environmental Sciences
6(1): 1-7, February 2022
e-ISSN:2622-0628
DOI:10.24843/ATBES.2022.v06.i01.p01
Available online at: https://ojs.unud.ac.id/index.php/ATBES/article/view/76081
Identification of Viral Nervous Necrosis in Grouper Fish (Epinephelus sp.) by Reverse Transcriptase-Polymerase Chain Reaction with Different RNA Extraction Methods
Yova Tresya Galingging 1*, Putu Eka Sudaryatma2, Pande Gde Sasmita Julyantoro1, Alfi Hermawati Waskita1, Artanti Tri Lestari2, Wahyu Nurlita2
1Department of Aquatic Resources Management, Faculty of Marine and Fisheries, Udayana University
Jl. Kampus Unud Bukit Jimbaran, Kuta Selatan, Badung, Bali 2Fish Quarantine and Inspection Agency Denpasar, Bali Jl. Sunset road No. 777, Kuta, Badung, Bali
*Corresponding author: yovagalingging@gmail.com
Abstract.ViralNervousNecrosis(VNN)isaNodaviridaevirusinfectedsnapperandgrouperfish,especiallyinthelarvaland seed stages targeted eye and nervous organ. Abnormal swimming was showed by fish behavior at the bottom pond. This study aimed to compared the identification of VNN through three different methods of RNA extraction from the organ target and amplification was carried out by Reverse Transcription of cDNA followed by Polymerase Chain Reaction (PCR) and quantitative real-time PCR (qPCR) technique. Herein, The RNA target from the organ from VNN-positive grouper fish samples targeted RNA2 region with 230-bp in electrophoresis gel and 69-bp by TAM-probe from qPCR were successfully converted and amplified from all RNA extraction kit. Different results of VNN amplified-cDNA was RNA extraction methods-dependent. Amplification using Tri Reagent® extraction kit showed the best result, with large amount of RNA genome with good purity and showed the clear results from PCR and qPCR. Furthermore, sensitivity of RNA extraction methods was different from 50-100 ng total RNA/reaction. It was concluded that all RNA extraction kits can be used for the detection of the VNN genome by PCR and qPCR technique while resulted in different of RNA yields. Overall, this study offers suitable methods to extract VNN genome from grouper fish.
Keywords: Comparison; PCR; RNA Extraction; VNN
-
I. INTRODUCTION
Fish as natural resource is existing and renewable in long time periods by aquaculture if followed by good management. As a renewable resource if this was not used properly, it can cause a decrease in the availability which can lead to extinction. To properly manage fish resources, scientific information is needed regarding fish biology, fish resource dynamics and the environment [1].
Grouper (Epinephelus sp.) aquaculture were good developed in Indonesia. It has high selling value both in the domestic and foreign markets. However, huge of problem of fish farmer to maintained stable culture. Fish diseases problems are often faced in aquaculture [2]. Viral Nervous Necrosis (VNN) is a Nodaviridae family that infected grouper fish. It can cause 100% mortality in the
larva and 30% death in adult fish less than 20 days of culture. This 20-25 nm virus is commonly replicated in the eye, brain and nervous organ systems with asymptomatic or exhibiting abnormal swimming behavior and stay at the bottom of the pond [3].
Asymptomatic of VNN-infected fish is the main source of transmission and potentially of spreading the diseases. To prevent the spreader of VNN between the farmers worldwide were needed to development of reliable diagnostic tests. The reverse transcriptase-polymerase chain reaction (PCR) based assays on fish organ is still considered the gold standard to detect VNN-infection. Definitely, PCR performance can be greatly affected by the efficiency of the viral RNA extraction procedures. Several methods are used in molecular biology to isolate RNA from samples, such as the guanidinium thiocyanate-
phenol-chloroform and anion-exchange resin extractions. Thus, many commercial kits exploit these methods to allow a fast, sensitive and reproducible detection of viral RNA. Therefore, our objective study is evaluating and optimizing the recovery of VNN RNA from VNN-positive fish organs using three different commercial kits as well as providing a suitable in-house extraction protocol. After methods optimization, the three methods of RNA extraction were evaluated and compared using different concentrations with RT-PCR for detection.
-
II. RESEARCH METHOD
-
A. Samples collection
The research was carried out for two months from December 2020 - January 2021 at the Laboratory of Fish Quarantine and Inspection Agency Denpasar branch (BKIPM Denpasar) Bali. Identification of this virus was used eyes and brain from juvenile stages of grouper VNN-positive. Each group of samples is amounted to three fish. All samples were stored in -20 ºC until used for RNA extraction. This research will be conducted using descriptive method. According to Sugiyono [4], descriptive research is used to analyze data by describing or describing the data that has been collected as it is without intending to make conclusions that apply to the public or generalizations. The location of the research was delivered in Fish Quarantine and Inspection Agency Regional Office Denpasar can be seen in Figure 1.

Figure 1. Research location in Fish Quarantine and Inspection Agency Regional Office Denpasar
-
B. RNA extraction
The principle of extraction is to separating nucleic acids from other cell components. According OIE [5] there are stages of extracted of genetic material by lysate of cell membrane, separated of genetic material from protein, and purified the of genetic material. In this study, we used three extraction methods to recovered VNN RNA from brain and eyes VNN-positive fish.
-
1. Tri Reagent® from SIGMA
Homogenize 20 mg of chopped sample with 500 µl of Tri Reagent® solution in the microtube. Then add 100 µl of Chloroform, vortex and kept at room temperature for 10 mins. Cold centrifugated at 12000 × rpm for 15 mins. Transfer 200 µl of supernatant into a new microtube which already contains 250 µl of 2-propanol and keep for other 10 mins. Again, centrifuge at 12000 × rpm for 15 mins and discard supernatant. Wash pellets by added 500 µl of 75% ethanol, and centrifuge at 7500 × rpm for 5 mins. Dry the pellet for 10 mins by air-drying. Dissolve the pellet with 200 µl of DEPC water. Isolated RNA was stored in -20 ºC until used.
-
2. IQ Plus™ Extraction Kit
Twenty mg chopped samples was homogenized with 500 µl Solution 1 in microtube. Add homogenate with 500 µl of Solution 2 and vortex. Then centrifuge 9000 × rpm for 1 min and transfer 500 µl of supernatant into a spincolumn tube. Continued centrifuge at 9000 × rpm for 1 min. Add another 500 µl solution 2 then centrifuge 9000 × rpm for 3 mins. Transfer the spin-column to microtube. Then add 200 µl of solution 3 and spin for 1 minute. Isolated RNA was stored in -20 ºC until used.
-
3. RNA Extraction® by GeneReach
Homogenate 20 mg of samples with 500 µl RNA Extraction solution and allowed to stand for 5 min at room temperature. Then, add 100 µl chloroform, vortex and kept at room temperature for 3 mins. Continued for centrifuged at 12000 × rpm for 15 mins. Transfer 200 µl of supernatant to microtube containing 200 µl of 2-propanol. Then centrifuge 12000 × rpm for 15 mins, discard supernatant. Wash pellet with 500 µl of 75% ethanol by centrifuged at 9000 × rpm for 5 mins, discarded ethanol and air-dried. Dissolve the pellet with 200 µl of DEPC water. Keep the isolated RNA in -20 ºC until used.
-
C. Reverse transcription and PCR condition
Amplification was divided into two procedures, prior to amplified the DNA, we converted RNA into cDNA using Reverse transcriptase, AMV (Promega, UK) following manufacturers protocol. The cDNA was assessed using a GoTaq® Green Master Mix (Promega, UK) to detect the RNA2 region of the VNN genome [6]. A conventional PCR method was used to amplification of cDNA, started with a PCR activation step at 94°C for 2 min followed by 30 cycles of denaturation at 94°C for 45 s, annealing 58°C for 45 s and extension at 72°C and extension temperature reached 72°C. Amplification of PCR products of RNA2 region target (230 bp) was confirmed by gel electrophoresis.
Quantitative real-time PCR (qPCR) was performed using a Rotor-Gene Q (Qiagen, USA) and QuantiTect® Probe RT-PCR (Qiagen, Germany) was used to detect the
VNN-positive gen from extracted organs. The specific primers and probe sequences (69 bp) were targeted RNA2 based on OIE [7]; their sequences were as follow forward primer: RNA2FOR: 5’-CAACTGACARCGAHCACAC-3’; reverse primer: RNA2REV: 5’-CCCACCAYTTGGCVAC; probe: 5’-FAM/TYTARGCRACTCGTGGTGCVG-3’.
Amplification condition was performed according to the manufacturer's specifications (Qiagen). Briefly, two microliters (μL) of cDNA was added to 11.5 μL of the reaction mixture. Each 11.5 μL reaction mixture contained 0.25 μL of each primer forward and reverse (10 μM), 9 μL of RNase-free Water, 0.75 μL of Real-time Buffer and 0.25 μL of QuantiTect RT enzyme. Reactions were incubated at 50 °C for 30 mins and 95 °C for 15 min followed by 40 cycles at 94 °C for 15 s and 55 °C for 45 s. All experiments were performed in duplicate and quantification cycle of threshold (Ct) values.
-
D. Electrophoresis
The amplified DNA was further analyzed using electrophoresis to be able to see whether the samples tested were positive or negative infected with the VNN virus. In principle, negatively charged DNA will move towards a
positive charge in an electric field. According to Seow [9] the displacement depends on the buffer conditions, temperature, duration of time, gel concentration and molecular size. The stages in the electrophoresis process are the manufacture of Agarose Gel, electrophoresis, and coloring.
-
E. Data analysis
Data analysis from gel electrophoresis was carried out descriptively obtained from the results by reading the results of the photo gel electrophoresis. According to Koesharyani [10], a sample that is declared positive for VNN if the sample band is at 230 bp in size or parallel to the band on the positive control. In the case of PCR results are declared negative result when the band undetected in the size of 230 bp or band was reached out from 230 bp size. On the other quantitative method, a qPCR experiments were performed in duplicate and quantification cycle of threshold (Ct) values that were below 40 were considered positive and declared to negative when Ct value above 40 (Qiagen).
-
III. RESULTS AND DISCUSSION
-
A. RNA isolated from three differences extraction methods
The RNA isolation method carried out in this study used several different methods so that the possible results of different RNA isolates [11]. The results obtained depend on the isolation technique used and the accuracy in the
work. This depends on the effectiveness of the method used in producing RNA isolates both in terms of quantity and quality as well as time efficiency in processing. Molecular techniques in the implementation to obtain data vary, both the technique and the desired target according to the facilities, human resources and funds [12].
The quality of the total RNA isolated was measured using a spectrophotometer to determine the concentration and absorbance ratio. The Tri Reagent® method is the highest method of producing RNA with a concentration value of 168.5. The total RNA concentration extracted using the RNA Extraction® method gave quite high results, between 43.5 - 86.5, while the lowest RNA concentration value was produced using the IQ Plus extraction method, which was 22 - 57.5.
The total isolated RNA concentration was measured using a spectrophotometer (Perkin Elmer, USA) at wavelengths of 260 and 280 nm. RNA concentration was measured at an absorbance of 260 nm with the calculation of every 1 absorbance value equal to 40 µg/mL [13]. The DNA purity from each method was calculated using 260/280 ratio. The highest purity was resulted from IQ Plus™ Extraction Kit method, while the lowest absorbance ratio was found in the Tri Reagent® method.
-
B. Evaluation of isolated RNA by Electrophoresis
The extraction method IQ PlusTM Extraction Kit is a new aquaculture diagnostic platform, which uses advanced insulated isothermal polymerase chain reaction technology to detect pathogen-specific nucleic acid sequences. The IQ Plus method also has a high level of sensitivity / high specificity and does not require a long time in the extraction process. The IQ Plus method can not only be used for disease diagnosis, but also used to detect at an early stage or subclinical contamination to prevent the spread of contamination.
1 2 3 4 5 6 7 8 9
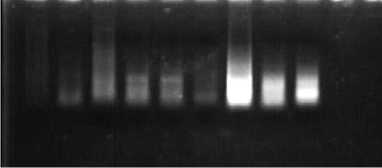
Figure 2. Direct electrophoresis to 1% Agarose gel of total isolated RNA from differences RNA extraction methods.
Information:
1st Well : Extraction Sample with IQ Plus 1
2nd Well : Extraction Sample with IQ Plus 2
3rd Well : Extraction Sample with IQ Plus 3
4th Well : Extraction Sample with RNA Extraction 1
5th Well : Extracted Sample with RNA Extraction 2
6th Well : Extraction Sample with RNA Extraction 3
7th Well : Extraction Sample with Tri Reagent® 1
8th Well : Extraction Sample with Tri Reagent® 2
9th Well : Extraction Sample with Tri Reagent® 3
The results of RNA isolation using the IQ Plus™ Extraction Kit method can produce good quality RNA as indicated by the presence of genomic RNA bands.
However, the results of electrophoresis showed that there was still a smear on the sample. The smear can be a remnant of the solutions that are still carried during the isolation process or can also be in the form of RNA that was degraded during the isolation process.
The RNA Extraction® method is a method of purification of RNA from samples of living organisms. Extraction of RNA in molecular biology experiments is very complicated in the presence of RNase and is strong which degrades RNA samples [14]. Certain RNases can be very potent and inactivating them is difficult compared to neutralizing DNases. This method is complicated by the presence of ribonuclease enzymes in cells and tissues, which can rapidly degrade RNA.
The results of RNA isolation using the RNA Extraction® method resulted in different purity values in each sample and different levels of RNA concentration obtained, this resulted in various band thicknesses and the presence of smears in the samples. The difference in RNA concentration obtained in each sample can be determined by the physical treatment carried out and the ability of the extraction buffer to break down cells. The process of physically destroying cells with perfect grinding of the sample can facilitate the extraction buffer in breaking up cells [15].
The Tri Reagent® extraction method is a mixture of a mixture of guanidine thiocyanate and phenol in monophase solution which is used to isolate DNA, RNA and protein from biological samples of humans, animals, plants, yeast, bacteria and viruses. The Tri Reagent® method is used to homogenize the biological samples from which RNA, DNA or proteins are extracted. The Tri Reagent® method is an improved version of the one-step total RNA isolation reagent developed by Rio [21]. This reagent-based RNA isolation method is widely used and proven for RNA applications. It is ideal for fast, economical and efficient total RNA isolation or simultaneous isolation of RNA, DNA and protein from human, animal, plant, yeast, bacterial and viral samples.
The results of RNA isolation using the Tri Reagent® method produced good quality RNA as indicated by the presence of genomic DNA bands. However, the results of electrophoresis are still possible smears due to incomplete homogenization or lysis of the sample and the final RNA pellet may not be completely dissolved.
-
C. Evaluation of differences RNA extraction methods by PCR
The results of RNA isolation using the three methods were then amplified to detect the presence of VNN from each method used. The success of virus detection using the PCR technique is determined by the provision of nucleic acid material with good concentration and purity. According to Li [16] high RNA concentration and low
number of inhibitors determine the success of the PCR technique in detecting pathogens.
1 2 3 4 5 6 7 8 9 10 11 12
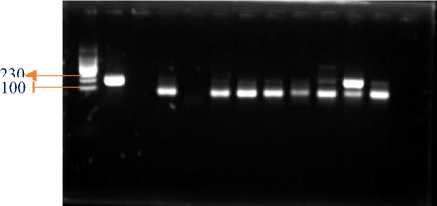
Figure 3. Electrophoresis of amplified PCR product from total isolated RNA
information:
1st Well : Marker
2nd Well : Positive Control
3rd well : Empty
4th Well : Extraction Sample with IQ Plus 1
5th Well : Extraction Sample with IQ Plus 2
6th Well : Extraction Sample with IQ Plus 3
7th Well : Extraction Sample with RNA Extraction 1
8th Well : Extraction Sample with RNA Extraction 2
9th Well : Extraction Sample with RNA Extraction 3
10th Well : Extraction Sample with Tri Reagent® 1
11th Well : Extraction Sample with Tri Reagent® 2
12th Well : Extraction Sample with Tri Reagent® 3
The results of electrophoresis show the success of RNA extraction using each extraction method, indicating that the extraction method used has succeeded in extracting RNA [15]. The DNA target in the grouper sample measuring 230 bp was successfully amplified. In each sample showed positive and negative results VNN using the three extraction methods.
-
D. Comparison on serial dilution of isolated RNA by
PCR and qPCR
Isolated RNA was performed again based on the modified protocols and analysed by both PCR and qPCR assays in comparison to those performed following manufacturer's instructions. Prior to the amplification process using PCR, DNA dilution was carried out first [17], where the amplification was carried to make an equality of RNA yield. The dilution is carried out in order to minimize the presence of contamination such as phenol, protein, and residual materials during extraction. In addition, dilution can also be used to determine the final volume of the RNA template in the composition of the PCR material.
The dilution was carried out based on the brightness of the visualization of the DNA bands based on the concentration results of the RNA extraction. The dilution results were then amplified according to the both PCR and qPCR. From adjusted RNA concentration in 50 ng/reaction, suggesting that extraction with Tri Reagent® extraction kit resulted in a clear band luminated (Figure 4b) and yielded the lowest Ct values in all three of samples (37.89; 19.55; 32.01; respectively). On the other hand, other two extraction methods were not resulting band in
PCR nor Ct value in qPCR in both concentrations. These results suggesting that Tri Reagent® extraction performance improves with viral RNA amount and clearly yieldofbandluminedandCtresultsfromPCRandqPCR, respectively.
1 2 3 4 5 6 7 8 9 10 11 12
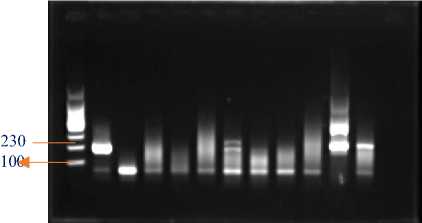
a. Dilution 100 ng/reaction
1 2 3 4 5 6 7 8 9 10 11 12
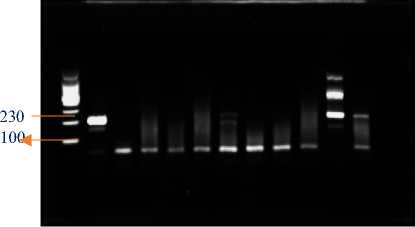
b. Dilution 50 ng/reaction
Figure 4. Electrophoresis of amplified PCR product from adjusted-concentration isolated RNA.
Information:
1st Well : Marker
2nd Well : Positive Control
3rd well : Empty
4th Well : Extraction Sample with IQ Plus 1
5th Well : Extraction Sample with IQ Plus 2
6th Well : Extraction Sample with IQ Plus 3
7th Well : Extraction Sample with RNA Extraction1
8th Well : Extraction Sample with RNA Extraction2
9th Well : Extraction Sample with RNA Extraction3
10th Well : Extraction Sample with Tri Reagent®® 1
11th Well : Extraction Sample with Tri Reagent®® 2
12th Well : Extraction Sample with Tri Reagent® 3
-
E. Comparison of RNA Extraction Methods
Extraction of RNA genome of grouper infected with VNN using three RNA extraction methods showed different results both in terms of quality and quantity of DNA produced. Comparison of RNA extraction can be seen in Table 2.
Based on the table above, the method that has fast time efficiency is the RNA extraction method using IQ Plus with a long processing time from the extraction stage to electrophoresis with a processing time of about three hours in an easy way. Then the RNA extraction method with the RNA Extraction method and also the Tri Reagent® method has almost the same processing time, which is about 6 hours of processing. But in terms of costs, the one that costs the most is the extraction method using the Tri Reagent® and IQ Plus™ methods, while the one that costs the least is using the RNA Extraction® method.
Of the three RNA extraction methods, the highest DNA concentration value was obtained from samples extracted using the Tri Reagent® method, which was 168.5 mg/ml. The lowest DNA concentration was obtained from samples extracted using the IQ Plus™ Extraction Kit method, which was 22 µg/ml. Then the RNA Extraction® method reached a concentration of 43.5 µg/ml. In this study, pure DNA isolates have not been found, because the value of DNA purity ranges from 1.8 to 2.0, while the pure DNA in this study only has a DNA purity value between 0.09380.6506.
The most sensitive method in isolating the genomic RNA of grouper infected with VNN is using the Tri Reagent® method. Because this method produces good RNA purity values with the highest RNA concentration. It is possible that the isolated VNN genome becomes larger in groupers with light infection levels. However, in terms of cost, it is less effective in detecting VNN attacks on groupers.
-
F. Discussion
Quantitatively, the concentration of RNA produced using the Tri Reagent® method resulted in a higher concentration compared to the IQ Plus™ Extraction Kit extraction method with RNA Extraction®, while the purity of the RNA produced almost had the same purity for the three methods. The extracted RNA was then amplified using PCR to produce 230 bp amplicons. The results show that the Tri Reagent® extraction method can be developed as an RNA preparation method for PCR detection which is quite simple, but needs to be a little expensive.
Virus identification using PCR technique can be successful if the supply of nucleic acid material with good RNA concentration and purity. Based on the results obtained, the RNA extraction method used the Tri Reagent® method with good RNA concentration and purity, that indicated identification of VNN gave positive results. Results Identification of VNN in grouper was carried out with a dilution of 50 ng/reaction and 100 ng/reaction still produced positive results in the PCR and qPCR from RNA extraction method with the Tri Reagent® kit.
According to Fitriatin [18], amplification is the main process in PCR. Amplification aims to increase the target RNA to be analyzed using agarose gel electrophoresis. One cycle in the amplification process includes four stages, namely pre-denaturation, denaturation, annealing and extension. In the amplification process can occur 30 to 40 cycles that can produce millions of RNA. According to Kurniawati [19] basically the amplification process that is propagated is DNA material. VNN viruses are RNA viruses, therefore, to convert RNA strands into doublestranded DNA, they must first be transcribed using the AMV reverse transcriptase enzyme. Reverse transcription
occurs at the pre-denaturation stage, which is the reverse transcription process, which is copying RNA into DNA, where single strands of RNA will be converted into cDNA.
According to Fitriatin [8], the RNA virus amplification process was possible used a two-step amplification method, firstly Reverse Transcriptase (RT) and continued with PCR. The amplification process was carried out in a PCR machine. The first stage of amplification is Reverse Transcriptase process is used to synthesize RNA into single-stranded complimentary DNA (cDNA) so that it can be amplified using a PCR machine. The main components in the PCR process are DNA templates, primers, dNTPs, Taq DNA polymerase and buffer solutions. According to Verkuil [20], nested RT-PCR makes it possible to reduce contamination of the product during amplification of unnecessary primer pooling. The nested primer will combine with the first PCR product and produce a shorter product than the first product. The amplified DNA was further analyzed using electrophoresis.
Identification of VNN virus in grouper based on the image resulted in 8 samples of grouper positive VNN and 1 sample of grouper negative VNN. But in the image the brightest band is found in well 11 with the Tri Reagent® extraction method. Positive samples with identified positive bands but with faint bands it is possible that the sample is contaminated, so amplification is carried out again by dilution and then electrophoresis is carried out using agarose gel.
Dilution of 100 ng/reaction resulted in 2 positive samples, namely in well 11 and well 12 where the method used in samples of wells 11 and 12 was the Tri Reagent® RNA extraction method. Dilution of 50 ng/reaction resulted in 1 positive sample, namely in well 11 using the RNA extraction method with the Tri Reagent® method. Of the three results of electrophoresis, the most effective and efficient method to identify VNN in grouper fish is using the extraction method, namely the Tri Reagent® extraction method.
-
IV. CONCLUSION
Identification of VNN genome from grouper’s organ using differences RNA extraction methods at BKIPM Denpasar was possible to resulting amplification curve in cycle threshold from qPCR results under 40 and luminated band from electrophoresis of all samples which was indicated by the of sized bands 230 bp and parallel to the DNA band in the positive control.
The RNA isolation method using three diferrences methods, namely IQ PusTM Extraction Kit, RNA Extraction® and Tri Reagent®, showed that the Tri Reagent® extraction method was an effective and efficient method in terms of time and process, but in terms of cost,
it was less effective in detecting VNN-positive in groupers fish.
REFERENCES
-
[1] Barani, H. M. 2012. Peranan Penting Ilmu Iktiologi dalam Kegiatan Usaha Penangkapan Ikan. Jurnal Iktiologi Indonesia, 2(2): 57-60.
-
[2] Adinapraja, D. 2015. Hama dan Pengendaliannya. Universitas Jendral Soedirman, Purwokerto.
-
[3] Yuasa, K., Koesharyani I., Roza D., Mahardika K., Johnny, F., dan Zafran. 2001. Manual for PCR Procedure: Rapid Diagnosis on Viral Nervous
Necrosis (VNN) in Grouper no.13. Lolitkanta – JICA. Gondol. p35- 38.
-
[4] Sugiyono. 2010. Metode Penelitian Pendidikan Pendekatan Kuantitatif, kualitatif, dan R & D. Bandung: Alfabeta
-
[5] OIE [Office International des Epizooties]. 2012.
Manual of Diagnotic Tests for Aquatic Animals Chapter 2.3.11, Viral Encephalopathy and Retinopathy (VER).
-
[6] OIE [Office International des Epizooties]. 2006. Newcastle disease. Dalam Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. World Org. Anim. Health, Paris, France.www.oie.int. Hlm. i–iii.
-
[7] OIE. 2010. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2010. www.oie.int
-
[8] Fitriatin E., dan Abdul M. 2015. Pemeriksaan Viral Nervous Necrosis (VNN) pada Ikan dengan Metode Polymerase Chain Reaction (PCR). Jurnal Ilmiah Perikanan dan Kelautan, Vol 7 (2) : 149-152.
-
[9] Seow, V. L., and A.R. Bahaman. 2012. Discriminatory Power of Agarose Gel Electrophoresis in DNA Fragments Analysis. In: Magdeldin S. Gel Electrophoresis – Principles and Basics. In Tech. Croatia. Hal 41-56.
-
[10] Koesharyani I, Novita H, 2006. Diagnosa Nested Reverse Transcriptase PCR untuk Viral Nervous Necrosis pada Benih Ikan Kerapu Bebek (Cromileptes altivelis). Journal Riset Akuakultur, 1(3): 33-40.
-
[11] Adiputra, J, Hidayat SH, Damayanti TA. 2012. Evaluasi tiga metode preparasi RNA total untuk deteksi Turnip mosaic potyvirus dari benih Brassica rappa dengan reverse transcription-polymerase chain reaction. Jurnal Fitopatologi Indonesia. 8(2):44-49.
-
[12] Ardiana. 2009. Teknik Isolasi DNA Genom Tanaman Pepaya dan Jeruk dengan Menggunakan Modifikasi buffer CTAB. Buletin Teknik Pertanian. 10: 11-13.
-
[13] Adriany, D.T., bakri, A.A., Bungalim, M.I. 2020. Perbandingan Metode Isolasi DNA Terhadap Nilai Kemurnian DNA untuk Pengujian White Spot
Syndrom Virus (WSSV) pada Lobster Bambu (Panulirus Versicolor). Prosiding Simposium
Nasional VII Kelautan dan Perikanan 2020 Fakultas Ilmu Kelautan dan Perikanan, Universitas Hasanuddin, Makassar, 5 Juni 2020
-
[14] Nurhayati, B., Darmawati, B., 2017. Biologi Sel dan Molekuler. Kementrian Kesehatan Republik Indonesia
-
[15] Mulyani, Y., Purwanto, A., Nurruhwati, I. 2011. Perbandingan Beberapa metode isolasi DNA Untuk DNA Untuk Deteksi Dini Koi Herpes Virus (KHR) Pada Ikan Mas (Cyprinus cario L.). J. Unpad. Ac. Id.
-
[16] Li R, Mock R, Huang Q, Abad J, Hartung J, Kinard G. 2008. A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogen. J Virol Methods. 154(2-3): 55-58.
-
[17] Ambarwati, A. 2018. Isolasi Dan Amplifikasi DNA Genomik Pasak Bumi (Eurycoma longifolia) Asal Sumatera Utara [Skripsi]. Medan: Fakultas Kehutanan Universitas Sumatera Uutara. 50 hlm.
-
[18] Fitriatin E., dan Abdul M. 2015. Pemeriksaan Viral Nervous Necrosis (VNN) pada Ikan dengan Metode Polymerase Chain Reaction (PCR). Jurnal Ilmiah Perikanan dan Kelautan, Vol 7 (2): 149-152.
-
[19] Kurniawati, M. D., Sumaryam., Hayati, N. 2019. Aplikasi Polymerase Chain Reaction (PCR) Konvensional Dan Real Time-PCR Untuk Deteksi Virus VNN (Viral Nervous Necrosis) Pada Ikan Kerapu Macan (Epinephelus Fuscoguttatus). Jurnal Techno-Fish. Vol 3 (1).
-
[20] Verkuil, E.V.A., van Belkum., dan J. P. Hays. 2008. Principles and Technical Aspects of PCR Amplification. Netherlands: Springer Science + Business Media B. V.
-
[21] Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. 2010. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor Protocols. Vol. June (6).
TABLE I
CONCENTRATION (µG ML-1) AND PURITY OF TOTAL RNA
|
Method |
Sample |
Wavelength |
Concentration (μg∕mi) |
Purity | |
|
260 |
280 | ||||
|
1 |
0,0115 |
0,0043 |
57,5 |
2,785 | |
|
IQ Plus™ Extraction Kit |
2 |
0,0449 |
0,0469 |
22 |
1,938 |
|
3 |
0,0102 |
0,00421 |
51 |
2,423 | |
|
1 |
0,087 |
0,0458 |
43,5 |
1,932 | |
|
RNA Extraction® |
2 |
0,093 |
0,0455 |
46,5 |
2,044 |
|
3 |
0,173 |
0,1534 |
86,5 |
2,324 | |
|
1 |
0,0322 |
0,0301 |
161 |
0,5358 | |
|
Tri Reagent®® |
2 |
0,0211 |
0,0259 |
105,5 |
0,3576 |
|
3 |
0,0337 |
0,0318 |
168,5 |
0,6506 | |
TABEL II
COMPARISON OF EFFECTIVENESS AND SENSITIVITY OF RNA
|
Method used |
Effectiveness of Detecting VNN | ||||
|
Time efficiency |
DNA concentration |
DNA Purity |
Amplification |
Cost | |
|
IQ Plus™ Extraction Kit |
+++ |
+ |
++ |
++ |
+ |
|
RNA Extraction® |
++ |
++ |
++ |
++ |
++ |
|
Tri Reagent®® |
++ |
+++ |
+++ |
+++ |
+ |
Information: + = Not good; ++ = OK; +++ = Very good
Discussion and feedback